Figure 5. Hourglass-shaped conservation of OCRs in mouse limb development.
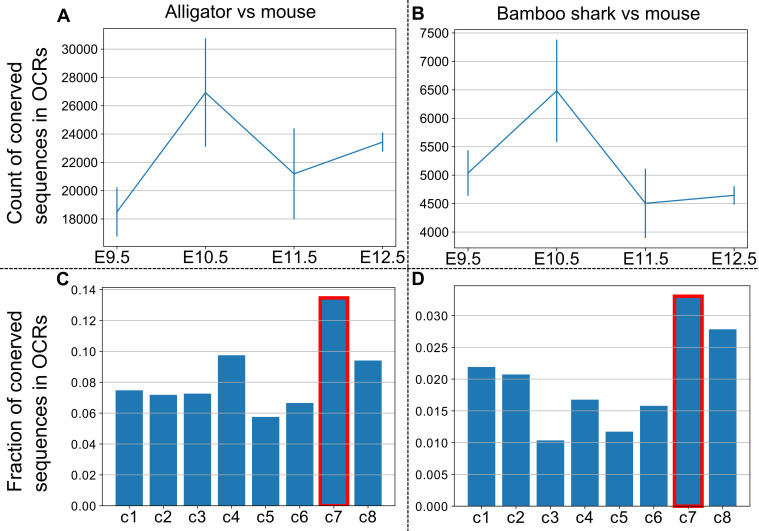
(A) ATAC-seq signals in the enhancer regions of the HoxA cluster. e1 to e4, known limb enhancers. Green vertical lines below the signals, peak regions determined by MACS2. (B) Comparison of a quality index, FRiP, for ATAC-seq data. Blue bars are samples with a FRiP score >0.2. The number in the end of the label name indicates the replicate number. (C) Conservation analysis of sequences in ATAC-seq peaks with BLASTN. The values to the right of each graph indicate the fraction of conserved sequences in the total peak regions. The common name of each genome sequence is indicated above the graph. The not-conserved heatmap indicates the fraction of sequences that were not aligned to any genome sequences and thus serve as a negative control. (D) Temporal changes of sequence conservation frequency in ATAC-seq peaks with LAST. Error bars: SEM.