FIGURE 1.
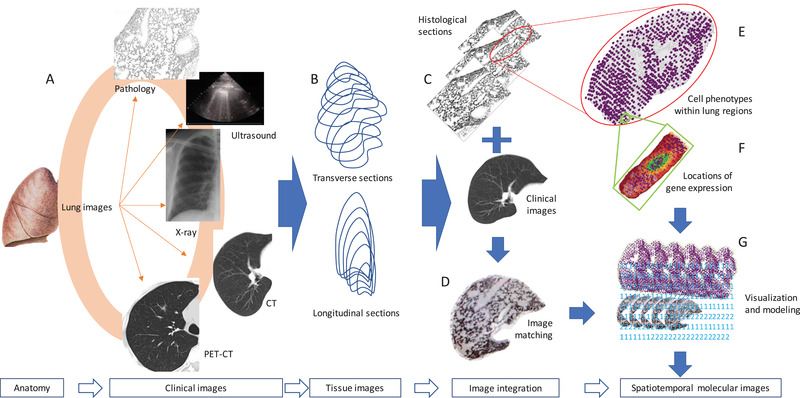
Proposed workflow of spatiotemporal molecular images. Clinical images of the organ anatomy are collected from e.g., X‐ray, computerized tomography (CT), nuclear magnetic resonance (NMR), positron emission tomography‐CT (PET‐CT), ultrasound, interventional radiology, and electrocardiogram (A). The three‐dimensional organ‐wide architectures and images can be re‐established by multiple transverse and longitudinal sections of tissues or clinical images (B). Histological sections of the organ are corresponded to clinical images (C) and then formed into an overlap by image matching (D), from which the histological structures of the organ are expected to be referred from clinical images. Spatial transcriptomes can simultaneously be performed on those histological sections in order to understand cell‐cell interactions, transcriptional signals, and phenotypes within the certain area (E) as well as mRNA expression within the cell (F). Multiple images at gene, cell, tissue, and organ levels are integrated into organ‐wide spatiotemporal molecular images using artificial intelligence, computerized programming and modeling, and visualization (G)
