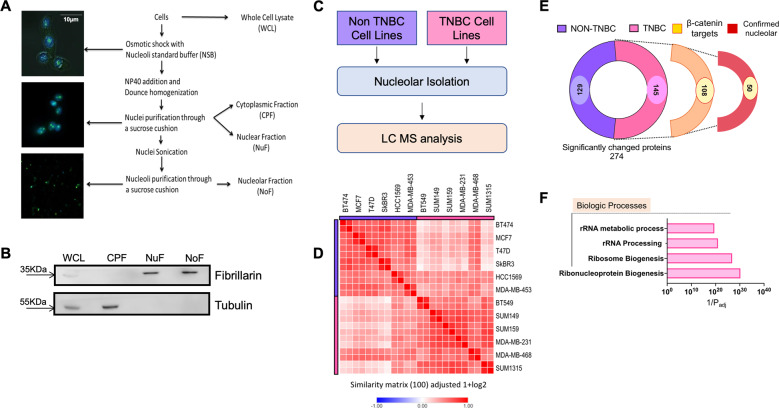
Fig. 2. Identification of differences in nucleolar proteomes of TNBC and non-TNBC cells.
A Graphical representation of the steps involved in isolating nucleoli from breast cancer cells. B Representative immunoblot showing verification of enrichment of nucleolar proteins. Fibrillarin is used as an indicator of nucleolar proteins. The absence of tubulin from the nuclear and nucleolar fractions was monitored to ensure purity. C Schematic summarizing workflow for nucleolar isolation and proteomic analysis to identify significantly altered proteins in the nucleolus between TNBC and non-TNBC cell lines. D Similarity matrix comparing nucleolar proteomes of TNBC cell lines with that of Non-TNBC cell lines. The proteomes were determined from two biological replicates. The matrix shows proteomes from both replicates of each cell line. E Graphical break-down of significantly altered proteins found in the nucleolar proteomics of the TNBC groups. Out of 145 proteins enriched in TNBC nucleoli, 108 were potential transcriptional targets of β-catenin. Out of these, 50 proteins were already reported to be nucleolar in one or more previously reported nucleolar proteomes from studies conducted in other (not breast cancer) cell types8,28,29. F Gene ontology analysis of proteins enriched in the nucleolar proteome of TNBC compared to Non-TNBC for biologic processes. The analysis was performed using g:Profiler™30.