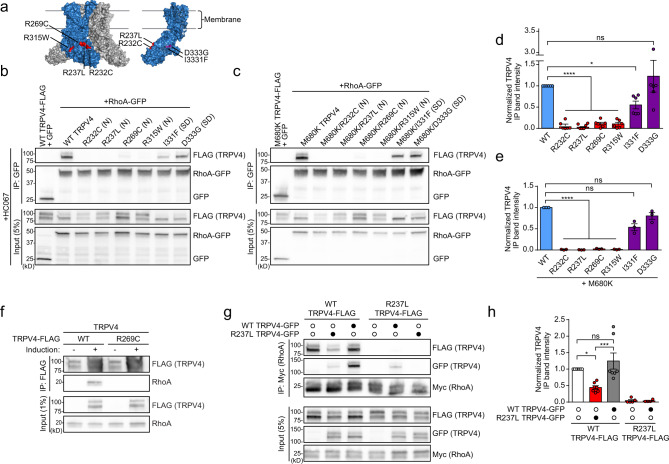
Fig. 2. TRPV4 neuropathy mutations disrupt RhoA interactions.
a Schematic showing the structure of the TRPV4 homotetramer (Xenopus tropicalis, PDB: 6BBJ) with a single subunit in blue. Neuropathy mutations (R232C, R237L, R269C, R315W) are highlighted in red, and skeletal dysplasia mutations (I331F, D333G) within the opposite face of the ARD (rotated for visualization) are highlighted in purple. b, c MN-1 cells were transfected with RhoA-GFP and TRPV4-FLAG with neuropathy-causing mutations (N) or skeletal dysplasia-causing mutations (SD) and treated with the TRPV4 antagonist HC067 (0.5 µM) (b) or RhoA-GFP and TRPV4-FLAG harboring an ion channel pore-inactivating mutation, M680K, either alone or in combination with neuropathy-causing mutations (N) or skeletal dysplasia-causing mutations (SD) (c). Cell lysates were then subjected to immunoprecipitation with anti-GFP antibody. Neuropathy mutations, but not skeletal dysplasia mutations, disrupt interaction between TRPV4 and RhoA, independent of calcium-mediated cytotoxicity. d, e Quantification of densitometry of immunoprecipitated TRPV4 band intensity divided by TRPV4 input band intensity, normalized to WT TRPV4 ((d); representative blot shown in b) or M680K TRPV4 ((e); representative blot shown in c). One-way ANOVA with Tukey post hoc test, n = 6 (b) and n = 3 (c) independent experiments, *p = 0.035, ****p < 0.0001. f Co-immunoprecipitation using T-Rex-TRPV4WT or T-Rex-TRPV4R269C cells demonstrates that WT but not neuropathy mutant TRPV4 interacts with endogenous RhoA. The presence of TRPV4-FLAG in uninduced immunoprecipitation lanes is due to low-level expression leak. g Co-immunoprecipitation of MN-1 cells transfected with TRPV4-FLAG and TRPV4-GFP constructs as well as RhoA-GFP demonstrates that neuropathy mutant TRPV4 reduces interaction of WT TRPV4 with RhoA (compare lane 1 vs. 2). h Densitometry of immunoprecipitated TRPV4 band intensity normalized to WT TRPV4 alone (lane 1); representative blot shown in g. One-way ANOVA followed by Tukey’s multiple comparisons test, n = 4 (lane 5), 5 (lane 4), or 7 (lanes 1, 2, and 3) independent experiments, *p = 0.028, ***p = 0.0009. Data are presented as mean ± SEM. Further details regarding the number of times experiments were repeated are presented in “Methods” under “Statistics and reproducibility”.