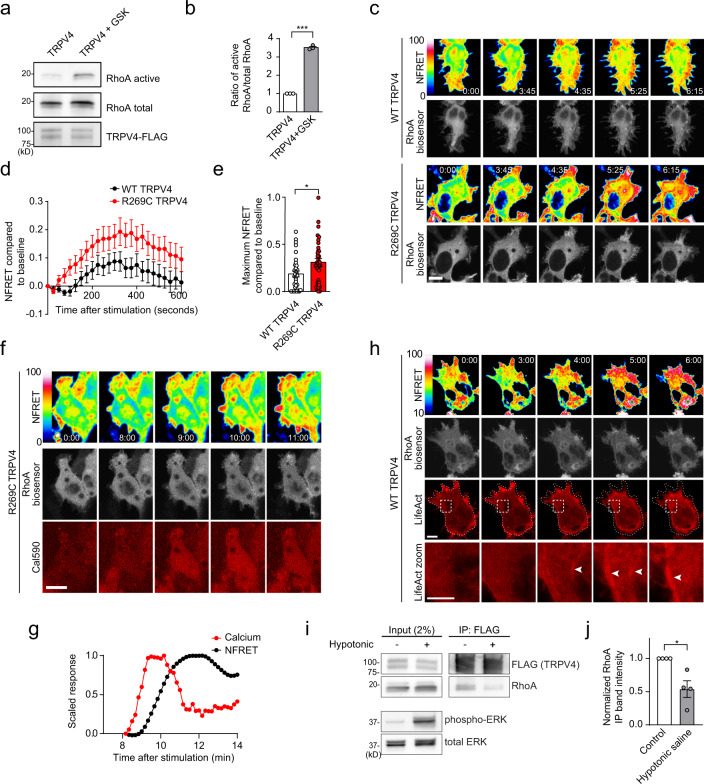
Fig. 5. TRPV4-mediated calcium influx activates RhoA and cytoskeletal changes.
a RhoA activation assay of T-Rex-TRPV4 cells induced with tetracycline (15 ng/ml, 16 h) in the presence of HC067, followed by removal of HC067 and treatment with GSK101 (5 min). b Densitometry of band intensities (active RhoA/total RhoA) normalized to untreated condition; representative blot shown in a. Paired two-tailed t-test, n = 3, ***p = 0.0007. c T-Rex-TRPV4 cells were transfected with RhoA2G biosensor and induced with tetracycline (15 ng/ml, 16 h) in the presence of HC067. Upon HC067 removal, cells were treated with GSK101 and RhoA FRET was assessed for 10 min. Confocal images show normalized FRET (FRET signal/FRET donor intensity) (top) and RhoA biosensor distribution (mVenus, bottom). Scale bars, 10 µm. d, e Averaged NFRET time course (d) and average peak NFRET (e) compared to baseline of GSK101-stimulated RhoA NFRET in cells expressing WT TRPV4 or neuropathy mutant TRPV4; representative images in c. Unpaired two-tailed t-test, n = 34 cells, *p = 0.0189. f, g Time series of a R269C TRPV4-FLAG-expressing cell showing changes in calcium (Cal590) and NFRET following treatment with GSK101 at time 0 (f); intracellular calcium and NFRET signals shown over time (g). Scale bar, 10 µm. h Time series of a WT TRPV4-FLAG-expressing cell co-transfected with LifeAct-mCherry and treated with GSK101 at time 0 showing actin stress fiber formation (arrowheads) and cellular contraction. Scale bars, 10 and 5 µm for inset. i T-Rex-TRPV4WT cells were induced in the presence of HC067 as in a followed by treatment with control media or hypotonic saline (5 min) followed by TRPV4 immunoprecipitation. Hypotonic saline causes partial disruption of TRPV4–RhoA interaction. Increased phospho-ERK (Thr202/Tyr204) indicates TRPV4-mediated calcium influx. j Densitometry of band intensities (immunoprecipitated RhoA/RhoA input); representative blot in i. Paired two-tailed t-test, n = 4, *p = 0.035. Data are presented as means ± SEM. Where indicated, HC067 was used at 0.5 µM; GSK101 at 100 nM. Further details regarding the number of times experiments were repeated are presented in “Methods” under “Statistics and reproducibility”.