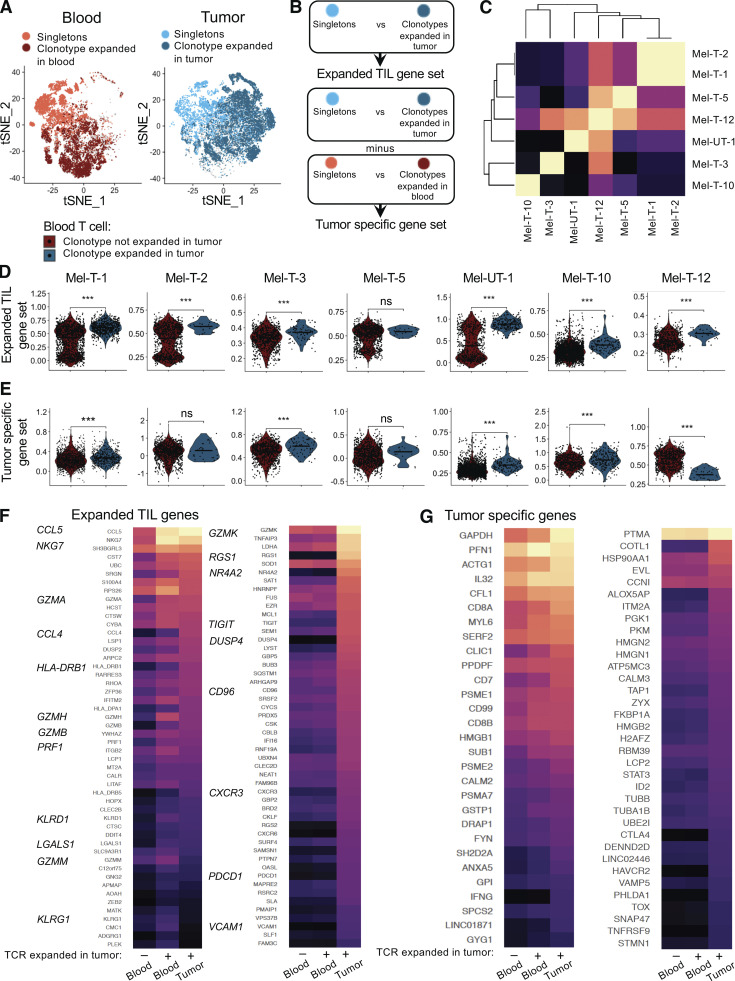
Figure 3.
Circulating TILs reflect features of clonally expanded tumor-infiltrating T cells. (A) tSNE plots highlighting tumor-infiltrating cells from 11 patients with a TCR clonally expanded in the tumor (dark blue) or singletons (light blue) and blood-derived cells with a TCR clonally expanded in blood (dark red) or singletons (tomato red). (B) Diagram illustrating the definition of expanded TIL gene sets and TIL-specific gene sets. (C) Clustered heatmap of the similarity matrix between expanded TIL gene sets of individual patients. (D) Quantification of the expression of expanded TIL gene sets in circulating TILs (blue) versus other circulating T cells (red) for each patient. Statistical significance was determined using Wilcoxon rank sum test (ns, P ≥ 0.05; ***, P < 0.001). (E) Quantification of the expression of tumor-specific gene sets in circulating TILs (blue) versus other circulating T cells (red) for each patient. Statistical significance was determined using the Wilcoxon rank sum test (ns, P ≥ 0.05; ***, P < 0.001). (F and G) For selected genes belonging to the expanded TIL gene sets of at least three patients, expression is compared between tumor-resident clonally expanded TILs, circulating TILs, and other circulating CD8 T cells. (F) Heatmap displaying expression of expanded TIL genes differentially expressed (left panel) or not (right panel) between circulating TILs and other circulating T cells from 11 patients. (G) Heatmap displaying expression of tumor-specific genes differentially expressed (left panel) or not (right panel) between circulating TILs and other circulating T cells from 11 patients. For heatmaps, values are scaled and mean-centered log2-transformed gene counts. Statistical significance was determined with Wilcoxon rank sum tests, Bonferroni-corrected by the number of genes in the dataset (17.045) multiplied by the number of comparisons (3) with a P value threshold of 0.05.