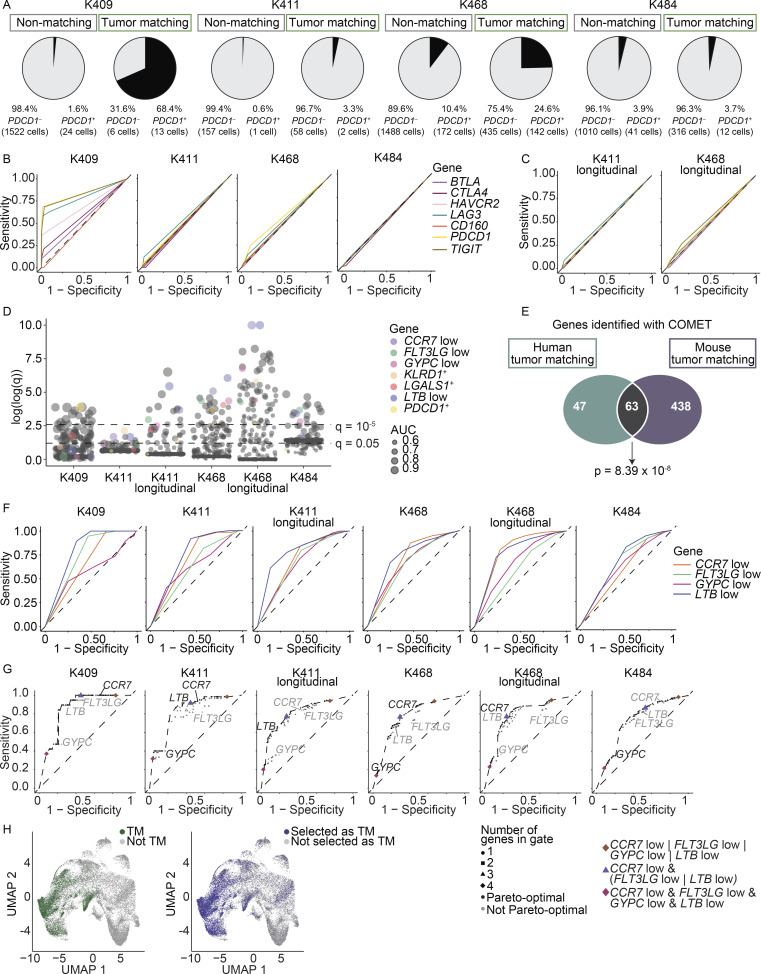
Figure 6.
Identification of combinations of markers for tracking TM cells across patients. (A) Frequency of PDCD1+ cells (using transcript) in the initial blood sample separated by TM and non-TM cells (four experiments). (B and C) ROC curves showing the sensitivity and specificity of inhibitory receptor transcripts to distinguish TM cells from non-TM cells in the initial blood samples (four experiments; B) and the longitudinal blood samples (two experiments; C). Legend shared between B and C. (D) Plot showing the significance values from the COMET analysis across blood samples. Significance using an XL-minimal hypergeometric test with multiple hypothesis test corrections. Circles sized by AUC for sorting TM cells from non-TM cells. The y axis corresponds to the log2(x + 1) transformation of the −log10 of the COMET q values, capped at 10. PDCD1 and consensus markers are highlighted with color. All other surface markers are gray. (E) Overlap between the single markers detected by COMET to distinguish TM cells from non-TM cells in the blood of mice (with MC38 tumors, M1–5 from three experiments) and patients (with melanoma, both treatment-naive samples and longitudinal samples, totaling six experiments). Markers included if detected as significant (q value < 0.05) in a minimum of two samples. Significance using a hypergeometric test, P = 8.39 × 10−8. Lists of genes and additional parameters in Table S11. (F) ROC curves for the consensus markers identified in D. (G) The sensitivity and specificity of all possible logic gates derived from combinations of genes CCR7low, LTBlow, GYPClow, and FLT3LGlow. Points are shaped by the number of markers used in the logical gate and colored black if they are pareto-optimal (if there is no gate with strictly better sensitivity and specificity) or gray if not pareto-optimal. A dotted line through the pareto-optimal gates represents the ROC of this combinatorial marker collection. (F and G) The dashed line represents the sensitivity and specificity values of random chance. (D, F, and G) Six experiments. (H) UMAP of CD8+ cells integrated from all patient blood samples (including longitudinal samples; data combined from six experiments). Left: True TM cells as defined by matching TCR sequence in green, nonmatching in gray. Right: Putative TM cells as determined by the best-performing gate, [CCR7low and (FLT3LGlow or LTBlow)], are colored blue; cells not expressing the marker combination in this gate in gray. For this combination, sensitivity = 0.780 and specificity = 0.716. The symbol “&” indicates the “and” gate, and the “|” indicates the “or” gate.