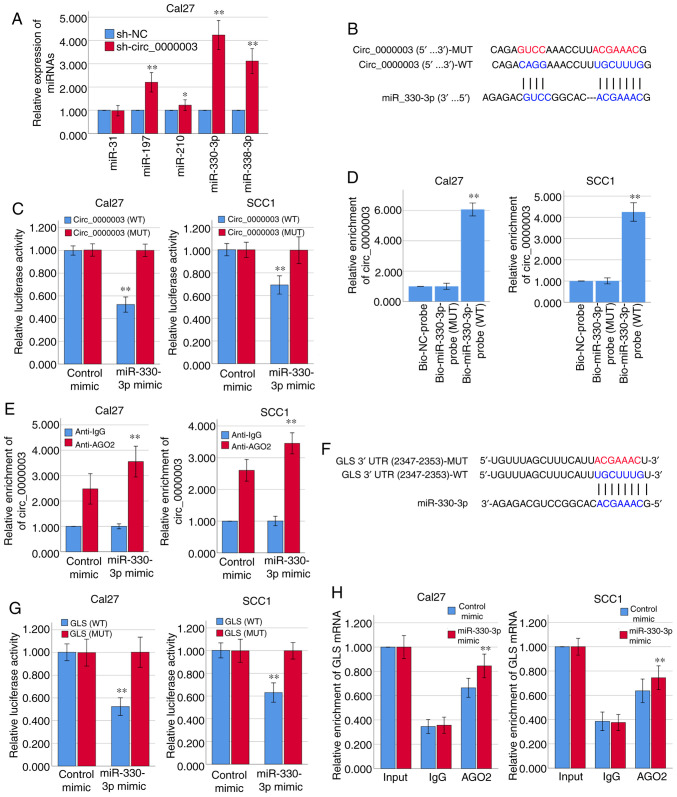
Figure 2.
Circ_0000003 modulates the tumor phenotype of TSCC cells via the miR-330-3p/GLS axis. (A) After transfection of sh-circ_0000003, expression levels of miR-31, miR-197, miR-210, miR-330-3p and miR-338-3p were detected via RT-qPCR. *P<0.05 and **P<0.01 vs. sh-NC. (B) Bioinformatics analysis suggested that miR-330-3p may share binding sites with circ_0000003. (C) Luciferase reporter assay indicated the molecular level combination of miR-330-3p and circ_0000003. **P<0.01 vs. circ_0000003(MUT). (D) Circ_0000003 was pulled down by biotinylated miR-330-3p-WT. **P<0.01 vs. Bio-miR-330-3p-probe(MUT). (E) AGO2-RIP followed by RT-qPCR to detect circ_0000003 enrichment level after miR-330-3p overexpression via miR-330-3p mimics. **P<0.01 vs. Anti-IgG. (F) Bioinformatics analysis suggested the associability within miR-330-3p and GLS. (G) Luciferase reporter assay indicated the covalent targeting of miR-330-3p with GLS mRNA 3′-UTR. **P<0.01 vs. GLS(MUT). (H) RIP assays using antibodies against AGO2 or IgG were performed in cellular lysates from Cal27 and SCC1 cells, and RT-qPCR demonstrated the relative enrichment of GLS mRNA in Cal27 and SCC1 cells transfected with miR-330-3p or control mimics. **P<0.01 vs. control mimic. circ_0000003, hsa_circ_0000003; TSCC, tongue squamous cell carcinoma; sh-circ_0000003, circ_0000003 shRNA; qRT-PCR, Reverse transcription and quantitative polymerase chain reaction; WT, wild-type; RIP, RNA immunoprecipitation; GLS, glutaminase; 3′-UTR, 3-untranslated region; MUT, mutant type.