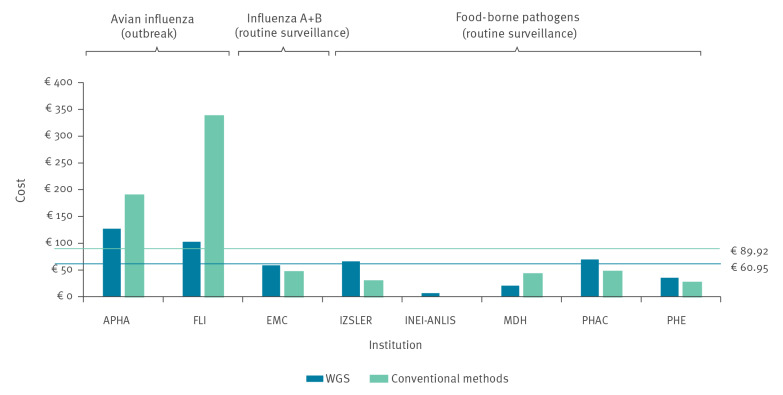
Figure 4.
Per-sample staff costs of whole genome sequencing vs conventional methods, case studies covering a specified reference period between 2016 and 2019 (n = 8 institutes)
APHA: Animal and Plant Health Agency; EMC: Erasmus Medical Centre; FLI: Friedrich-Loeffler-Institut; INEI-ANLIS: Instituto Nacional de Enfermedades Infecciosas - Administración Nacional de Laboratorios e Institutos de Salud; IZSLER: Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia-Romagna; MDH: Maryland Department of Health; PHAC: Public Health Agency Canada; PHE: Public Health England; WGS: whole genome sequencing.
Blue line: average costs of WGS; green line: average costs of conventional methods. For INEI-ANLIS, breakdown by cost type was not possible for conventional methods. FLI used a non-routine method as comparator (sequencing of a whole genome of a virus using Sanger sequencing). In contrast, APHA used the more limited and less resource-intensive haemagglutinin/neuraminidase analysis, a more typical conventional method for routine analysis.