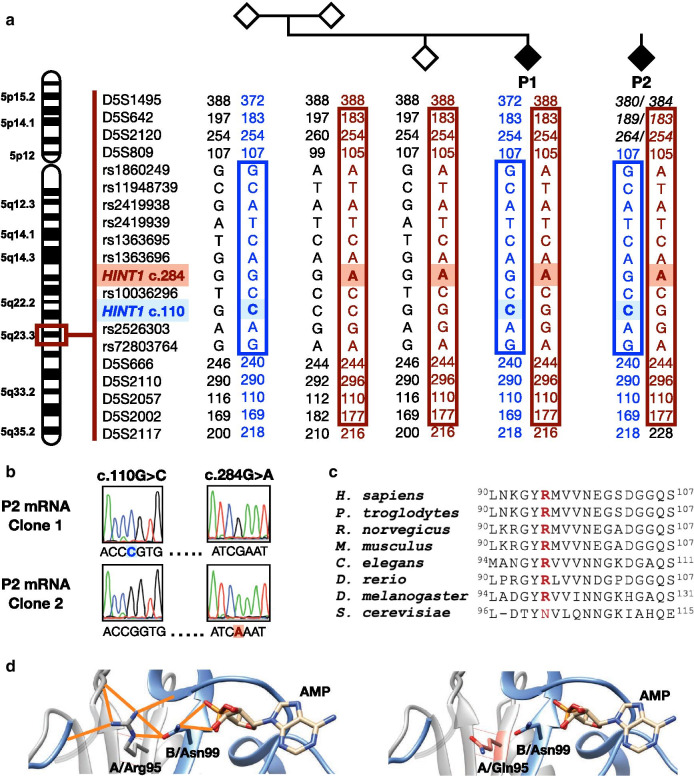
Fig. 1.
Genetic and in silico characterization of the identified variants in HINT1. a Pedigree structure, segregation and haplotype analysis. The shared haplotype surrounding the novel c.284G > A (p.Arg95Gln) variant is depicted in red, the previously established haplotype surrounding the known c.110G > C (p.Arg37Pro) founder variant is in blue. b Phasing analysis of the two HINT1 variants found in P2 using cDNA obtained from lymphoblasts. Electropherograms of two individual cDNA clones show that the variants are situated on different transcripts (in trans). c Evolutionary conservation of the amino acid residue affected by the novel p.Arg95Gln substitution in HINT1. The mutated residue is indicated in red. d Left: Localization of the p.Arg95 residue on the crystal structure of HINT1 (PDB:5KLZ) and its interaction with other amino acids through hydrogen bonds (orange). Right: The p.Arg95Gln substitution (in salmon) causes a loss of hydrogen bonds