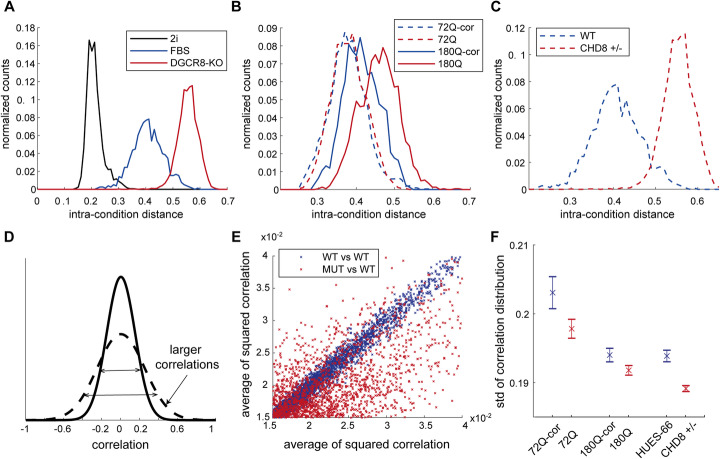
Fig. 2.
Cellular models of neurological disorders show increased transcriptional heterogeneity. a Heterogeneity of cell populations measured by “intra-condition distance” (see Methods) in mouse ESCs comparing DGCR8-KO cells (red) and WT cells grown in either 2i (black) or serum (blue) media conditions. b Same as a for human NPCs derived from two HD isogenic pairs: mutant 72Q cells (dashed red) and their isogenic controls (dashed blue); and mutant 180Q (solid red) and their isogenic controls (solid blue). c Same as a for human NPCs derived from WT hESCs (blue) and CHD8+/− isogenic cells (red). d A model for transcriptional disarray resulting in weaker correlations throughout the GRN. Shown is the distribution of correlations between all pairs of genes in the more organized state (dashed line) and the transcriptionally out-of-balance state (straight line). e The average of squared correlation of a gene with the rest of the genes. Each dot represents a single gene. Red, 180Q mutant vs corrected. Blue, two random pools of WT cells (see Methods). f Standard deviations of gene correlations in WT vs mutant cells in the 72Q, 180Q, and CHD8+/− isogenic systems. Error bars represents 1 std. over 10 replicates (see Additional file 1: Fig. S4 and Supplementary Methods)