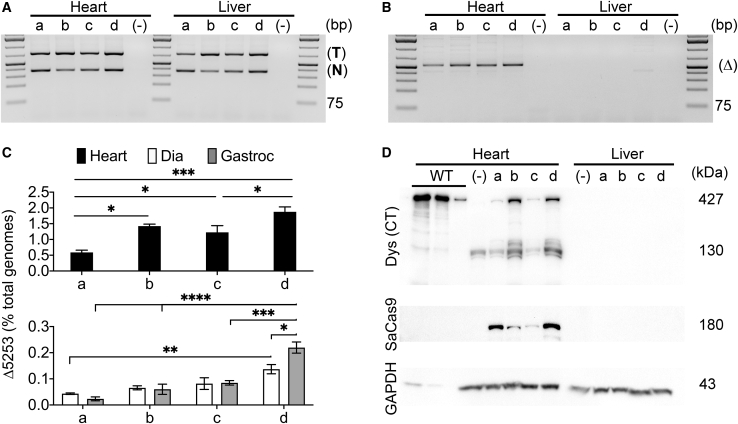
Figure 3.
Systemic Dystrophin Correction Is Enhanced with Increased Vector Dose and Demonstrates Preferred Dependence on Target versus Nuclease Vector Availability
Comparison of dose and ratio of muscle-specific nuclease and target vectors (Figure 1A) at 12 weeks following systemic delivery into young-adult (11-week-old mdx4cv) mice. The dose and ratios are the same as in Figure 2. (Top two lines) Semiquantitative PCR depicting the presence of nuclease (367 bp product) and target (741 bp product) vgs (A) or the dystrophin exons 52-53 (Δ5253) genomic DNA-deletion product (Δ; 520 bp product) (B) in DNA isolated from hearts and livers of mdx4cv mice treated with varying doses of nuclease/targeting vectors: (a) 1 × 1013/2 × 1012 vgs, (b) 2 × 1012/1 × 1013 vgs, (c) 5 × 1012/5 × 1012 vgs, and (d) 1 × 1013/1 × 1013 vgs; (−), untreated. PCR amplicon size reference via GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific). (C) Quantitative dPCR analysis of exons 52-53 (Δ5253)-deleted genomes versus total genomes for the various nuclease and target vector doses (a–d), as described above. (Top) Values for the heart; (bottom) diaphragm and gastrocnemius (Gastroc) values. Statistical significance was determined by two-way ANOVA with Tukey’s post hoc test. Values represent mean ± SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). (D) Western analysis of cardiac versus liver lysates for dystrophin (detected with a C-terminal dystrophin antibody [Dys (CT)]) and SaCas9 expression. The results show dose-dependent and muscle-specific expression of near-full-length dystrophin in hearts but not livers of treated mice. Lower molecular weight bands observed in cardiac samples following C-terminal dystrophin antibody staining likely reflect partial dystrophin degradation and/or possibly shorter CRISPR-induced dystrophin isoforms of unknown therapeutic relevance. A wild-type (WT; C57BL/6) cardiac muscle control sample was loaded at 10%, 5%, and 1% of total protein loaded for treated and untreated mdx4cv mice.