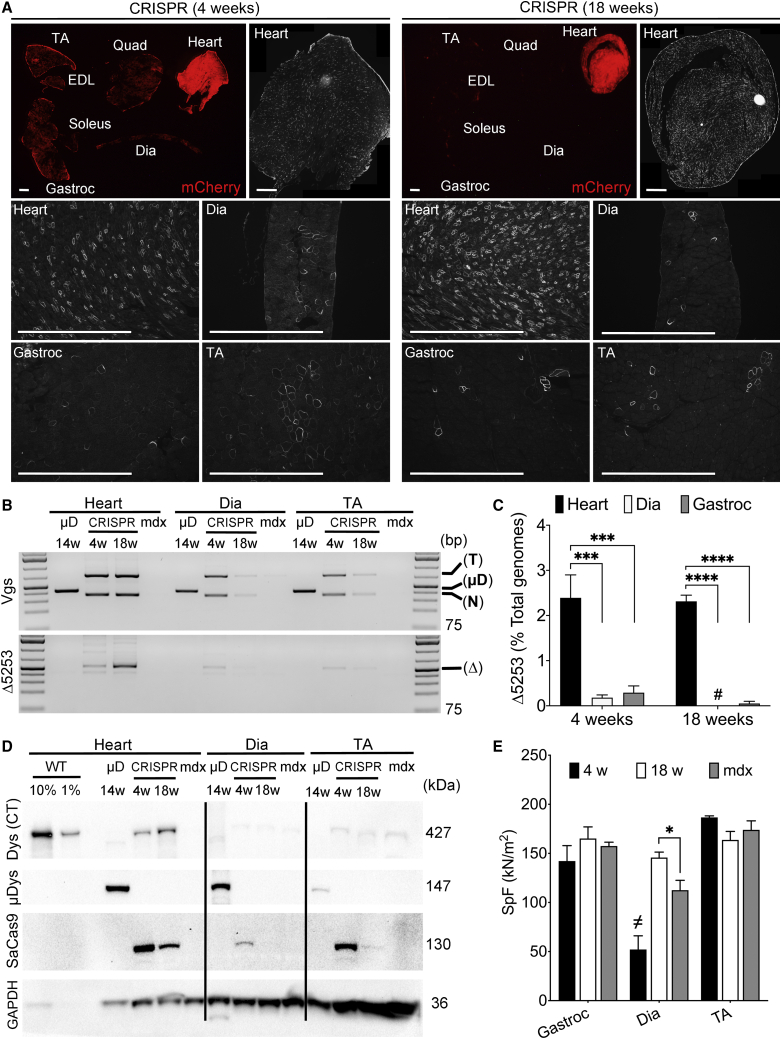
Figure 4.
Dystrophin Correction Persists in the Heart but Is Lost over Time in Skeletal Muscles following Gene Editing of Exons 52-53 in 2-Week-Old mdx4cv Mice
(A) Muscle transduction levels, as detected by expression of mCherry from target vectors at 4 and 18 weeks post-transduction (top) is reduced in all muscle groups except the heart between 4 (left) and 18 (right) weeks post-CRISPR treatment. Whole heart cross-sections (assembled from multiple 100× magnification images) show widespread dystrophin expression in cardiac myocytes at both 4 and 18 weeks post-treatment (top rows, right). (Bottom) At either 4 or 18 weeks post-treatment, abundant dystrophin-expressing myocytes in hearts but only rare dystrophin-positive fibers in skeletal muscle groups, such as diaphragm, gastrocnemius, and TA. Scale bars, 1 mm. (B) Semiquantitative PCR of vgs (top) or edited genomic DNA (Δ5253; bottom) in mdx4cv mice systemically treated with AAV6-CK8e-μDys5 (μD) or AAV6-CRISPR-SaCas9 (CRISPR). Mice were infused with vector at 2 weeks of age and analyzed 14 weeks later (μD) or 4 and 18 weeks later (CRISPR). The PCR product sizes are the following: μDys vector (μD; 429 bp), target vector (741 bp), and nuclease (SaCas9) vector (367 bp) (top). The PCR product from exon 52-53-deleted genomes (Δ5253 or Δ) is 520 bp (bottom). PCR products from untreated mdx4cv muscle extracts are shown as a control. The GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific) was included as a size standard. (C) Quantification via dPCR shows significantly enhanced Δ5253 correction in hearts versus diaphragm and gastrocnemius muscles at either 4 or 18 weeks post-transduction. Whereas cardiac Δ5253 correction remains stable between 4 and 18 weeks post-treatment, corrected genomes in diaphragm and gastrocnemius muscles appear to be drastically reduced during this same time period (also see Table 1). Statistical significance determined by two-way ANOVA with Tukey’s post hoc test. Values represent mean ± SEM (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Values that were too low to be visualized on the graphs are demarcated by # (see Table 1). (D) Western analysis of muscle lysates derived from heart, diaphragm, and TA muscles of CRISPR- or μDys5-treated mice showing expression of near-full-length dystrophin (Dys (CT)), μDys5 (μDys), SaCas9, and GAPDH. The results show a loss of edited dystrophin expression between 4 and 18 weeks in skeletal muscles but an increase in expression in the heart, despite lower SaCas9 expression at the later time point in all tissues examined. Conversely, μDys expression is clearly detected at 14 weeks post-transduction in all muscle groups. (E) Functional assessment of specific force generation in multiple skeletal muscle groups of CRISPR-treated (4 weeks [w], 18 w) versus age-matched untreated control (mdx) mice, including gastrocnemius, diaphragm, and TA muscles. Whereas only minor improvements were observed for diaphragm at 18 weeks post-treatment, there are no significant improvements in treated gastrocnemius or TA muscles at any time point when treatment is initiated prior to the rapid skeletal muscle turnover phase in mdx mice. The significantly reduced values in specific force for diaphragm at the 4-week time point (demarcated as ≠) are related to the immature state of the diaphragm at this early stage, making relevant comparisons challenging.23 Statistical significance determined by two-way ANOVA with Tukey’s post hoc test. Values represent mean ± SEM (∗p < 0.05).