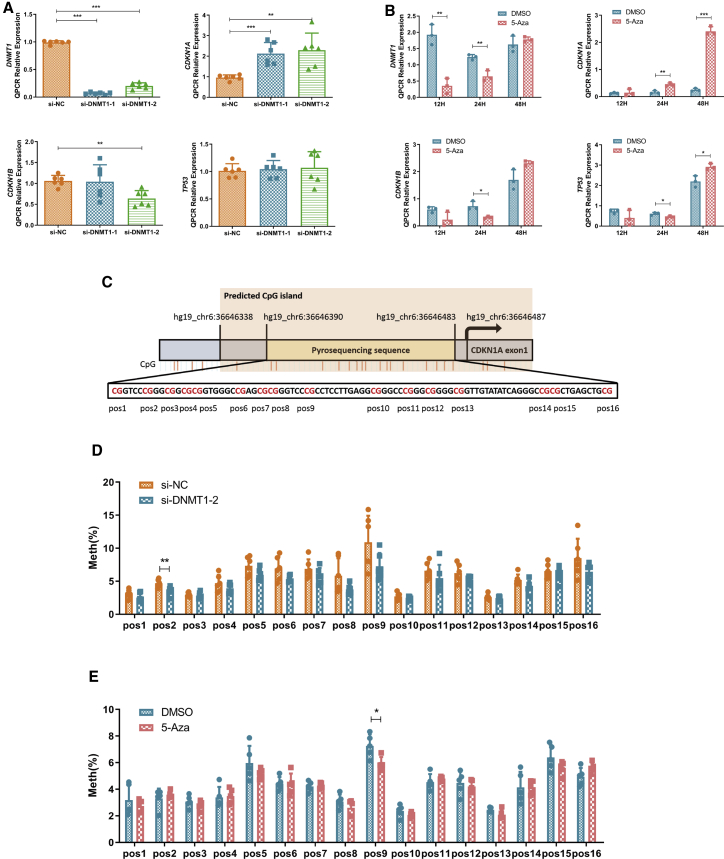
Figure 5.
DNMT1 Silencing or 5-Aza Treatment Upregulated CDKN1A Expression by Decreasing Its Promoter Methylation Levels
(A and B) Quantitative real-time PCR analysis of DNMT1 and negative proliferation marker levels (CDKN1A, CDKN1B, TP53) after DNMT1 silencing using two different siRNAs (A, n = 6 for each group; DNMT1, p < 0.0001; CDKN1A: si-NC versus si-DNMT1-1, p = 0.0004; si-NC versus si-DNMT1-2, p = 0.003; CDKN1B: si-NC versus si-DNMT1-2, p = 0.0012) or after 5-Aza treatment (5 μM) for different durations (B, n = 3 for each group; DNMT1: 12 h, p = 0.0022, 24 h, p = 0.0057; CDKN1A: 24 h, p = 0.0035, 48 h, p < 0.0001; CDKN1B: 24 h, p = 0.0145; TP53: 24 h, p = 0.0324, 48 h, p = 0.0179). (C) The selected pyrosequencing sequence was located upstream of CDKN1A exon 1 and had 16 CpG sites named pos1–pos16. CpG island enrichment in the CDKN1A promoter region was predicted by the MethPrimer website, which was located in the promoter and first exon regions of CDKN1A. (D and E) Methylation status of the CDKN1A promoter CpG island after DNMT1 silencing using two different siRNAs (D, n = 6, pos2, p = 0.0073) or after 5-Aza treatment (5 μM) for different durations (E, n = 5, pos9, p = 0.0166). Pos, CpG site in the CDKN1A promoter; Meth, methylation level. Mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test).