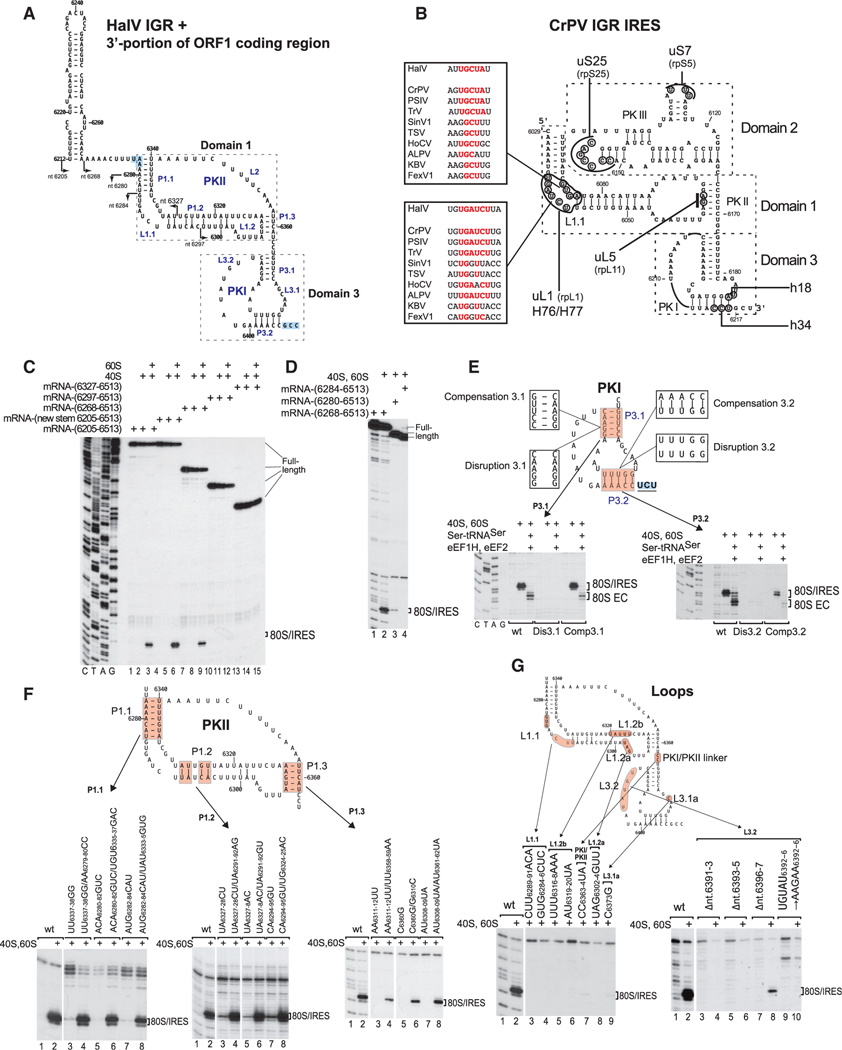
Figure 3. Structure and Mutational Analysis of the HalV IGR IRES.
(A) Model of the HalV IGR IRES and the adjacent 3'-terminal region of ORF1, derived on the basis of computational analysis (see STAR Methods) and chemical/ enzymatic foot-printing (Figures S1A and S1B). It is annotated to show nucleotides at 20 nt intervals, IRES domains and secondary structural elements (based on the nomenclature proposed by Costantino and Kieft (2005)), the ORF1 stop codon (UAA6276) and the ORF2 A site codon (GCC6406) (both boxed blue). Arrows indicate the 5′ borders of truncated HalV IGR mRNAs used in experiments to assay IRES activity in ribosome binding (C and D).
(B) Model of the CrPV IGR IRES, showing IRES domains and secondary structure elements, nucleotides that interact with ribosomal proteins and elements of 18Sand 28S rRNAs, and conserved motifs in the L1.1 loop in dicistrovirus IGR IRESs (Nishiyama et al., 2003; Pfingsten et al., 2006).
(C and D) The 5′-terminal border of the HalV IGR IRES assayed by toe-printing of IRES/80S complex formation. The position of IRES/80S binary complexes is indicated. Lanes C, T, A, and G depict HalV sequence.
(E and F) Analysis of the influence of disruptive and compensatory substitutions in helical elements of domain 3/PKI (E) and domain 1/PKII (F) on the HalV IGR IRES’s ribosome-binding (E and F) and elongation (E) activity, analyzed by toe-printing. The positions of IRES/80S binary complexes and 80S ECs are indicated.
(G) Analysis of the influence of substitutions in single-stranded elements of the HalV IGR IRES on its ribosome-binding activity, analyzed by toe-printing. The position of IRES/80S binary complexes is indicated. (F and G) Separation of lanes by white lines indicates that they were juxtaposed from the same gel.