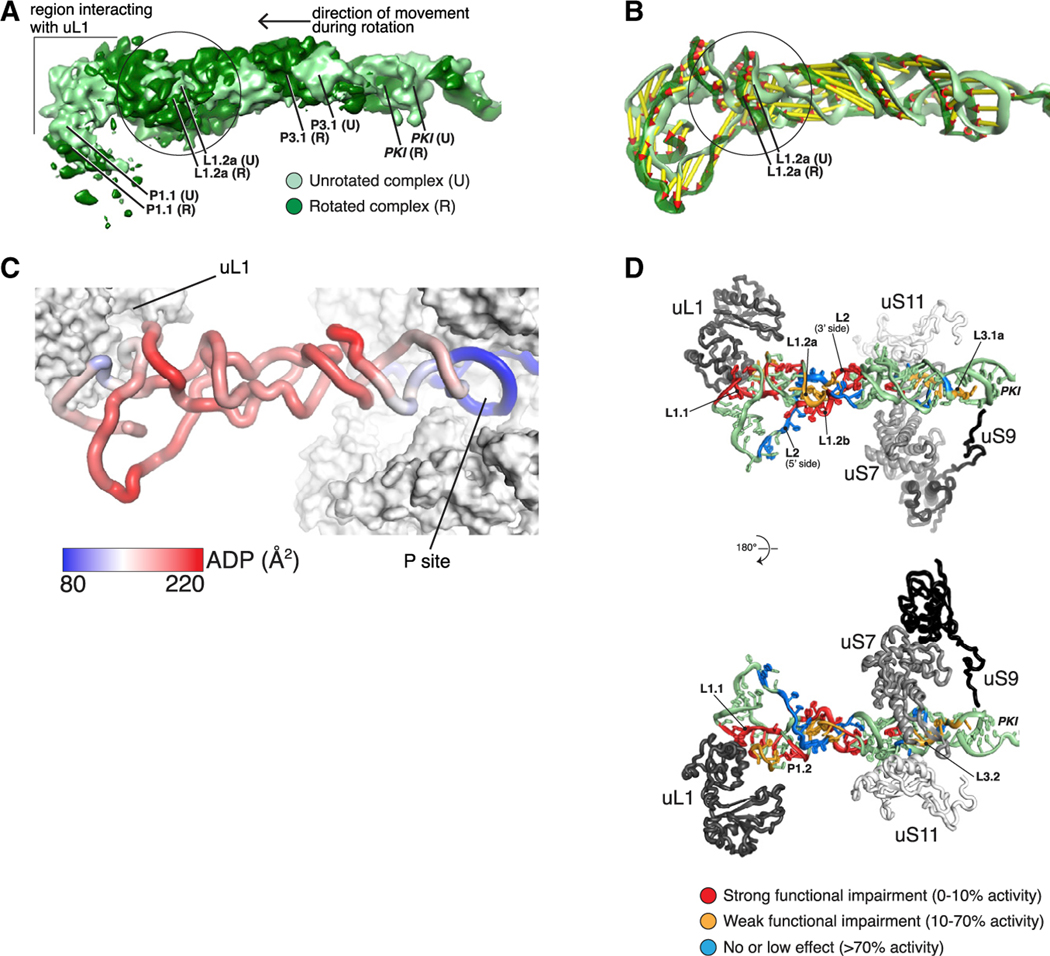
Figure 5. Flexibility of the HalV IRES Is Key to Its Function.
(A) Compression of the IRES in its central region during rotation. Superimposition of filtered density maps (Gaussian filter 1.5) of the unrotated and rotated states of the 80S-bound IRES. The circle indicates the region where additional bulging density is observed in the rotated complex.
(B) Superimposition as in (A) of the IRES 3D models (color coding as in A). Vectors help visualize the direction of the movement as well as its amplitude. Vectors were calculated by measuring the distance between phosphate atoms.
(C) The central region of the IRES is the most dynamic. Color coding of the unrotated IRES by atomic displacement parameter (ADP).
(D) Mutations that impair function cluster to regions interacting with ribosomal proteins and to the central region. Color coding of the IRES according to the effect of mutations on its function. Percentage of activity in comparison with wild-type RNA.