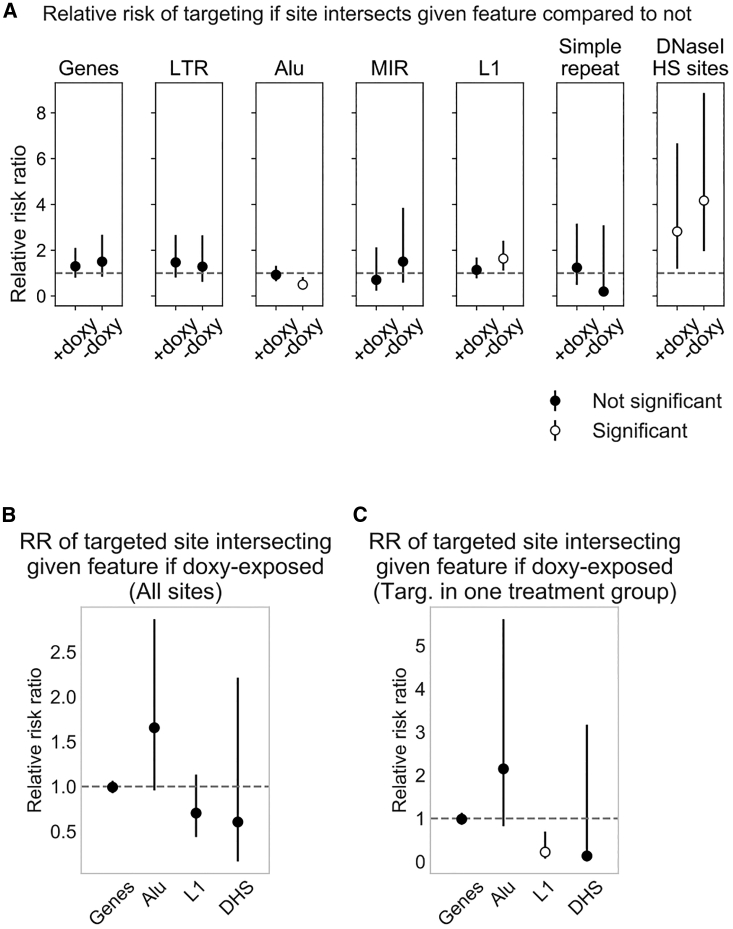
Figure 4.
Association of Targeted Sites with Chromosomal Features
(A) Relative risk ratio of a site having at least one integrated barcode, given that the site intersects the feature indicated above the plot. Sites were assembled into a 2 × 2 contingency table for which the exposure is intersection/no intersection and the outcome is targeted/not targeted. There were no 0 values in the tables. +doxycycline, n = 2,013; −doxycycline, n = 1,888. 95% confidence intervals are shown. We consider the relative risk ratio statistically significant when the 95% confidence interval does not overlap 1, shown by white circles. DNase I hypersensitive sites were obtained by intersecting provirus sites with DNase I-seq called peaks (see Materials and Methods). Low-complexity repeats were excluded due to large confidence intervals. (B and C) Relative risk ratio of a targeted site intersecting the feature indicated, given that it was targeted in a doxycycline-treated sample, for (B) all sites (+doxycycline, n = 181; −doxycycline, n = 137) or (C) sites targeted in only one treatment group (+doxycycline, n = 64; −doxycycline, n = 25). For (C), provirus sites were initially filtered to sites present in both treatment groups and targeted exclusively in one treatment group, as in Figure 3 (Group-specific). For (B) and (C), only targeted sites were assembled into a 2 × 2 contingency table for which the exposure is +doxycycline/−doxycycline and the outcome is intersection/no intersection. 0.5 was added to all cells for tables with a 0 value, as was the case for DNase I hypersensitive sites in (C). DHS, DNase I hypersensitive site.