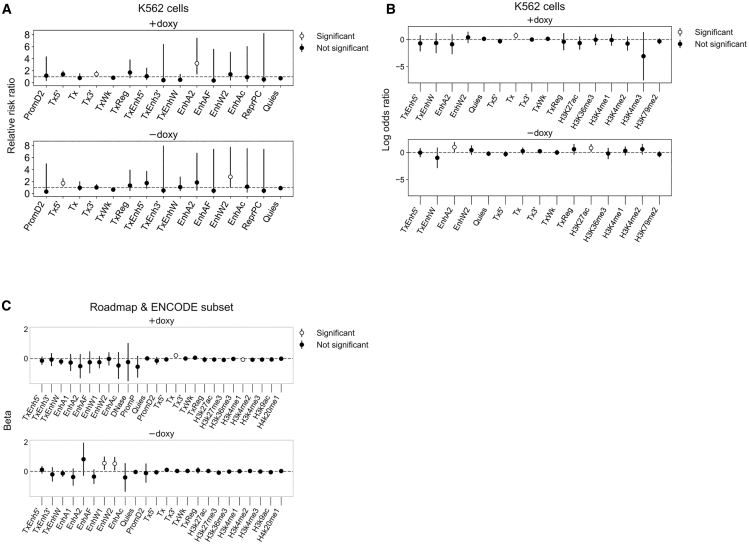
Figure 6.
Chromatin States and Epigenetic Measures Associated with AAV-HR
(A) Relative risk ratio of a site having at least one integrated barcode, given that it overlaps the indicated ChromHMM chromatin state segment, using chromatin state predictions in K562 cells. Sites were assembled into a 2 × 2 contingency table for which the exposure is intersection/no intersection and the outcome is targeted/not targeted. 95% confidence intervals are shown. We consider the relative risk ratio statistically significant when the 95% confidence interval does not overlap 1, shown by white circles. States with large confidence intervals were excluded but are given in Tables S4 and S5. Where incidence for either group is 0, 0.5 was added to all cells prior to computing relative risk ratio. (B) Predicting the presence of an overlapping ChromHMM or ENCODE feature peak in K562 cells from barcode heterogeneity at targeted sites using independent logistic regression models, filtering out features with high standard deviation. Exp(log odds ratio) represents the change in odds of overlapping a given feature for every unit increase in barcode heterogeneity. (C) Predicting barcode heterogeneity at targeted sites from the proportion of cell types assigned to a given ChromHMM state or ENCODE feature peak in the region over that site using independent linear regression models, filtering out features with high standard deviation. The cell types are a subset of seven cell types shared by both the Roadmap and ENCODE annotations (GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, and NHLF). Beta represents the mean change in barcode heterogeneity given a one-unit change in the proportion of assigned cell types. For (B) and (C), data were centered and scaled to a mean of 0 and standard deviation of 1 prior to model fitting. 95% confidence intervals are shown. A feature is considered predictive of targeting when the 95% confidence interval does not overlap 0, shown by white circles. Estimates and standard errors for all states are provided in Tables S6 and S7. For regression analyses, sites with barcode heterogeneity greater than the third quartile+3 × IQR of their respective treatment group were excluded.