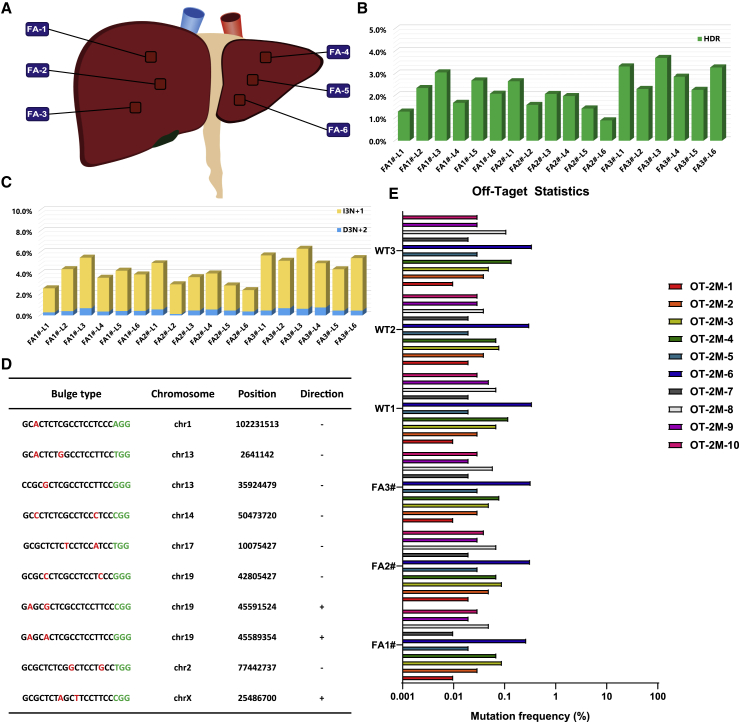
Figure 4.
CRISPR-Cas9-Mediated In Vivo Genome Editing Partially Corrects FAH Mutation in the Livers of HT1 Rabbits without Off-Target Effects
(A) Schematic views of the six positions of the HT1 rabbit livers injected with AAV (three for the right lobe and three for the left lobe) were collected for deep sequencing. (B and C) Histogram of the gene-corrected patterns, including precise HDR (B) and out-of-frame to in-frame mutations (3N+1 bp insertion and 3N+2 bp deletion) (C). (D) List of the top 10 potential off-target sites for sgRNA4. Mismatched nucleotides are indicated in red, and PAM sequences are labeled in green. (E) Targeted deep sequencing results for the top 10 potential off-target sites in the livers of the wild-type rabbits and AAV-treated HT1 rabbits.