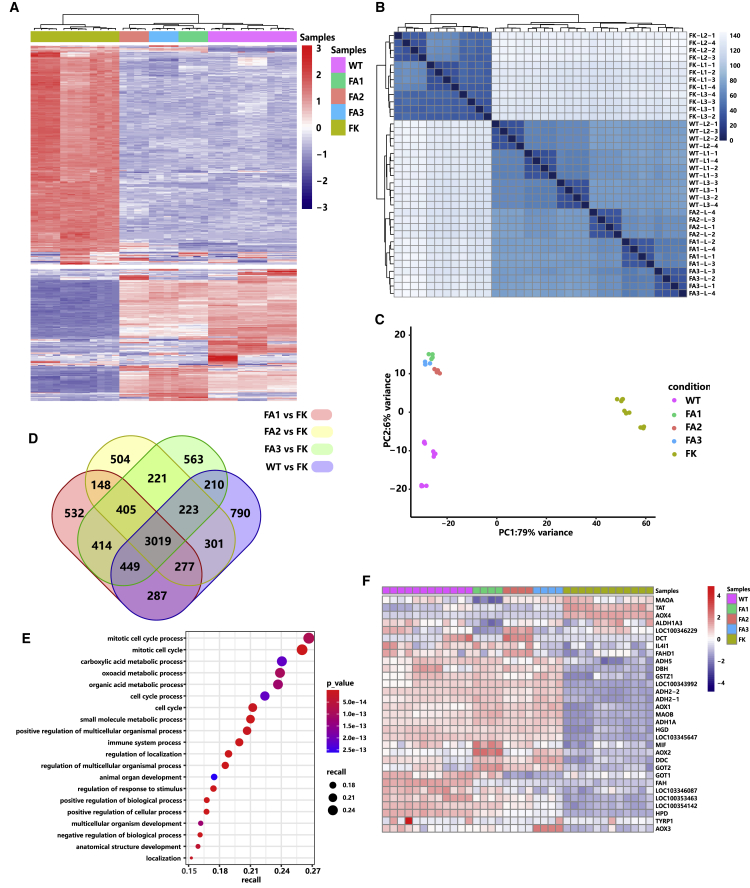
Figure 5.
Global Transcriptome Patterns of the Gene-Corrected HT1 Rabbits
(A) Hierarchical cluster analysis of the top 500 highest variance genes shows the transcriptome differences in the livers of the AAV-treated HT1 rabbits (FA1, FA2, and FA3), the PBS-treated HT1 rabbits (n = 3), and the wild-type rabbits (n = 3). Red and blue color intensities represent gene upregulation and downregulation, respectively. (B) Sample-to-sample distance analysis of the livers of the wild-type (n = 3), AAV-treated (n = 3), and PBS-treated (n = 3) HT1 rabbits. (C) Principal-component analysis of the wild-type rabbits (n = 3), AAV-treated HT1 rabbits (n = 3), and PBS-treated HT1 rabbits (n = 3). Points with different colors represent various sample groups. The percentage of variability explained by each component is shown on the axis. (D) Number of differentially expressed genes in the livers of the AAV-treated HT1 rabbits (n = 3) and the wild-type rabbits (n = 3) compared with the AAV-untreated HT1 rabbits (n = 3) (Venn diagram). (E) Gene Ontology (GO) enrichment analysis of the 3,019 commonly differentially expressed genes in (D). (F) Heatmap of the relative expression levels of genes related to the tyrosine metabolism pathway between wild-type rabbits (n = 3), AAV-treated HT1 rabbits (n = 3), and PBS-treated HT1 rabbits (n = 3).