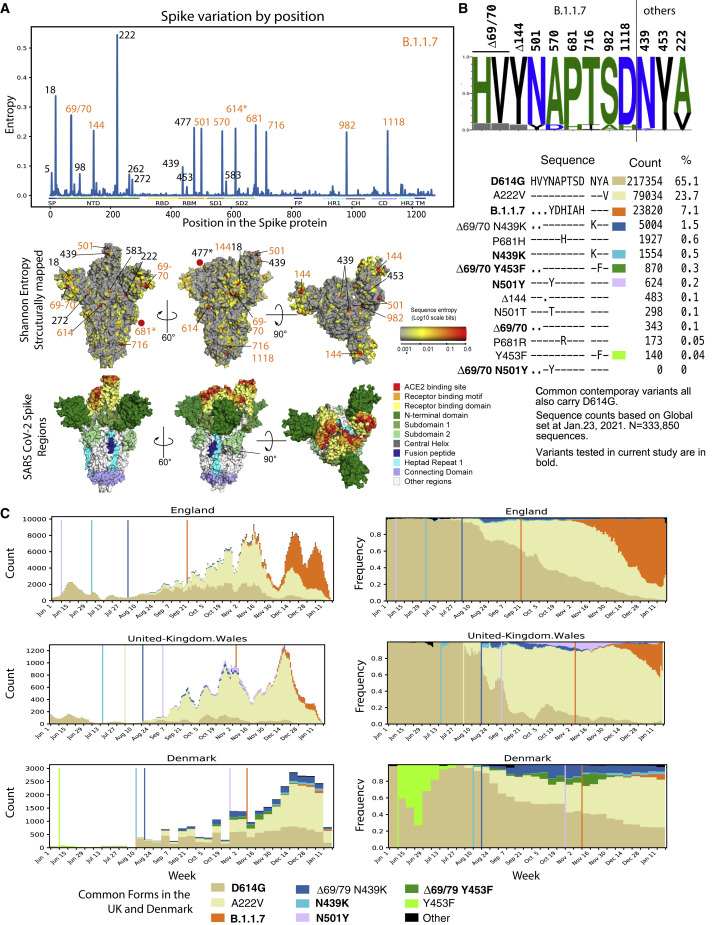
Figure 1.
Epidemiology tracing of mutations in B.1.1.7 and co-circulating relevant mutations in the UK and Danish SARS-CoV-2 epidemics
(A) Entropy scores summarizing the level of diversity found in positions in spike. These scores are dependent on sampling, and recent sampling from the UK and Denmark has been particularly intense relative to other regions of the world (Figure S1). B.1.1.7 mutations are highlighted in orange. The subset of B.1.1.7 sites with greater entropy scores (69/70, 681, and 501) are also often found in the context of other variants. The most variable site in spike is at 222 and is indicative of the GV clade. G614 has dominated global sampling since June 2020, and the entropy at 614 reflects presence of the ancestral form, D614, sampled in the early months of the pandemic. These same entropy scores are first mapped by linear position in the protein and then mapped onto the spike structure below the graph. Regions of spike are indicated by the same colors in the linear and structural maps.
(B) Frequencies of variants in relevant positions. Using the Analyze Align (AA) tool at cov.lanl.gov, we extracted the columns of interest for the B.117 spike mutations, and the additional sites of interest at 439, 453, and 222, out of a 333,850-sequence set extracted from GISAID on January 23, 2021. The logo at the top indicates the AA frequency in the full dataset; the gray boxes indicate deletions. All common forms of combinations of mutations at these sites of interest are shown, followed by their count and percentage. The forms that were common in the UK and Denmark are each assigned a color and used to map transition in frequencies of these forms over time in (C).
(C) Weekly running averages for each of the major variants in the UK and Denmark, based on the variants shown in (B), are plotted; the actual counts are on the left, and relative frequencies on the right. Some windows in time are very poorly sampled, some very richly. The vertical lines indicate when a variant is first sampled in a region. Note the lavender N501Y in Wales; this is N501Y found out of the context of B.1.1.7 and transient. The shifts in relative prevalence from the G clade (beige, D614G) to the GV clade (cream, A222V) to the B.1.1.7 variants (orange) are shown.