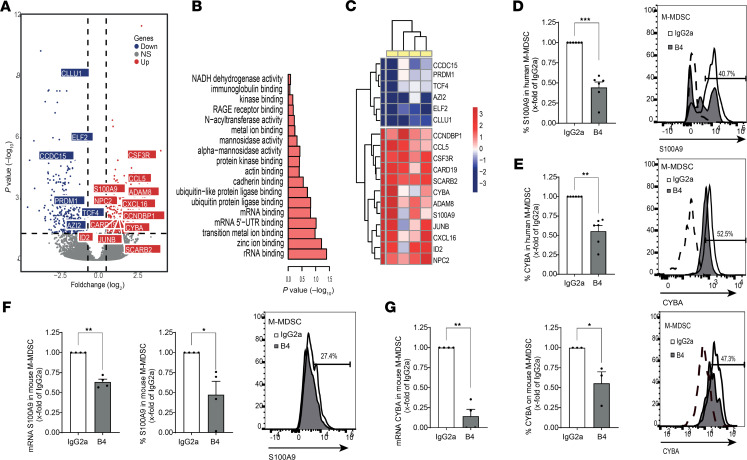
Figure 8. CD84 regulates functional and immunosuppressive pathways in human MM M-MDSCs.
(A–C) Sorted BM M-MDSCs (CD14+, CD15–, HLA-DR–, CD11B+) were activated with anti-CD84 or -IgG1k antibodies. After 24 hours, RNA was purified and sequenced using MARS-seq. (A) Volcano plot for M-MDSC genes, with gray indicating non-DE genes and with DE-expressed genes shown either in blue, for downregulated, or red, for upregulated (DE genes had P < 0.05 after adjustment and a base mean of more than 5 after adjustment with the R package, fdrtool; n = 4). (B) Highlighted GO terms from the upregulated DE genes of M-MDSCs (Molecular Function, EnrichR). (C) Heatmap depicting highlighted genes from the DE genes, with each column representing the log2 CD84 activated to IgG control ratio value of a single patient (n = 4). (D and E) Purified primary MM BM samples were incubated with B4 or IgG2a antibodies. After 48 hours, M-MDSCs were analyzed for S100A9 (D) (n = 6) or CYBA (E) protein expression (n = 6). (F and G) Sorted primary M-MDSCs (message) or whole BM cultures (protein) from 5TGM1-injected mice were incubated with B4 or control antibodies. After 24 hours (mRNA, left) or 48 hours (protein, right), cells were analyzed for S100A9 (F) and CYBA (G) expression. Representative histograms are shown (n = 3–4). One-tailed Student’s t test was used for F and G.