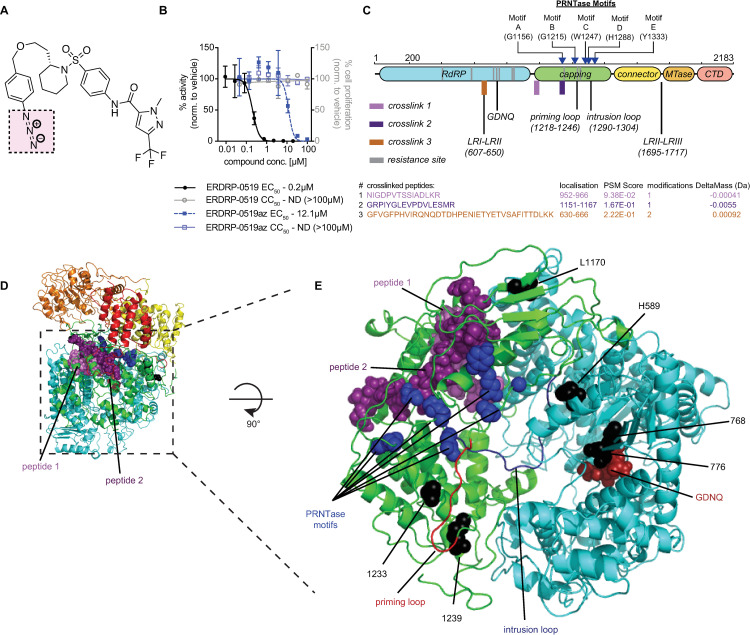
Fig 5. Photoaffinity labeling-based target mapping of ERDRP-0519.
A) Structure of photoactivatable compound ERDRP-0519az; the reactive azide moiety is highlighted (pink square). B) ERDRP-0519az is bioactive and displays no appreciable cytotoxicity. Symbols show means of three biological repeats, graphs represent 4-parameter variable slope regression models where possible. EC50 and CC50 are shown. C) 2D-schematic of the MeV L protein with locations of crosslinked peptides identified by photoaffinity labeling (top). The RdRP (cyan), capping (green), connector (yellow), MTase (orange), and C-terminal (light-red) domains, locations of known unstructured regions (LRI-LRII; LRII-LR-III), the GDNQ active site, specific amino acids motif in the capping domain, and positions of the intrusion and priming loops are shown. Sequences of peptides engaged by ERDRP-0519az (bottom). Specified are peptide location, spectrum match (PSM) score, number of ligands present, and delta mass. D) Location of peptides 1 (pink) and 2 (purple) in MeV L. E) Close-up top view of the capping and RdRP domains showing the adjacent locations of peptides 1 and 2. The priming loop (red), intrusion loop (blue), PRNTase motifs (blue spheres) and GDNQ active site (red spheres) are shown.