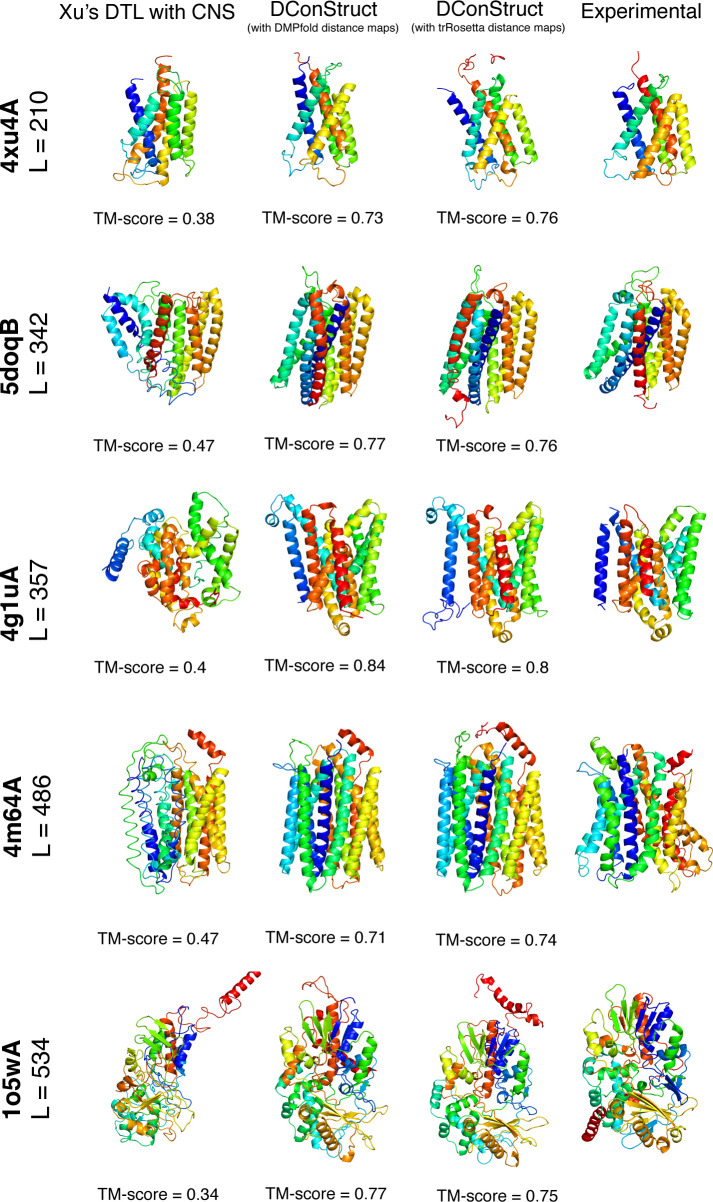
Fig 7. Ribbon diagrams of 3D models of MPs predicted using Xu’s DTL with CNS and DConStruct (using DMPfold- and trRosetta-predicted distance maps) along with the experimental structures for five protein targets: PDB ID 4xu4 and chain A, PDB ID 5doq and chain B, PDB ID 4g1u and chain A, PDB ID 4m64 and chain A, and PDB ID 1o5w and chain A.
All molecules are rainbow-colored blue to red from the N- to C-termini. Models are optimally superimposed on the experimental structures and then separated by translations along the horizontal direction.