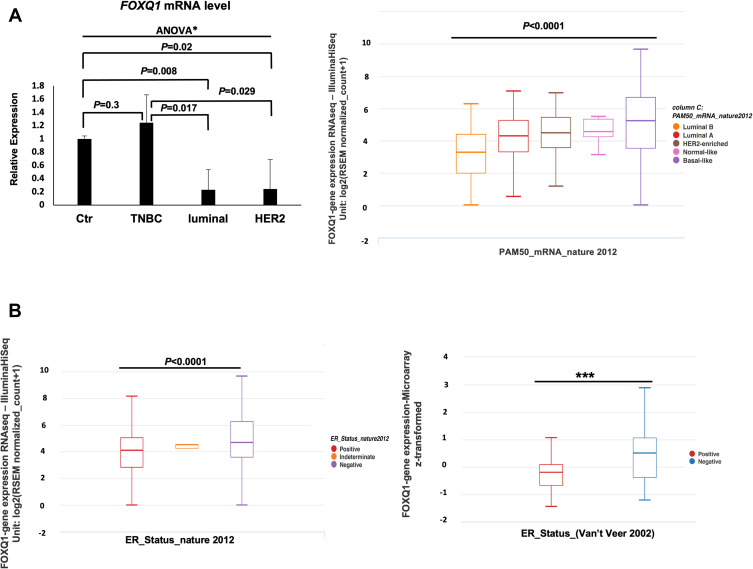
Figure 1.
FOXQ1 is differentially expressed across breast cancer subtypes and is under-expressed in luminal and HER2 breast cancer patient samples. (A) Left, qPCR experiments were conducted to measure FOXQ1 mRNA levels in normal breast tissue (Ctr, n=6) acquired from reduction mammoplasties, TNBC (n=6), luminal (n=6), and HER2 (n=6) BC patient samples. One-way ANOVA analysis was used to asses statistical difference in all groups *F(30,52) = 3.8, p=0.026 followed by unpaired t-tests and Bonferroni corrections were used for multiple comparisons. α= 0.05, adjusted α= 0.0125. The ΔΔCT method was used for analysis and TFRC was used as a reference gene. Error bars represent standard error of the mean (SEM). (A) Right, box plots showing FOXQ1 expression across BC intrinsic subtypes (PAM50). FOXQ1 RNAseq data in BC subtypes from the TCGA-BRCA data95 were used. Luminal B (orange), n = 127, Minimum = 0.00, Q1 = 2.0, Median = 3.29, Q3 = 4.40, Maximum = 6.26; Luminal A (red), n = 231, Minimum = 0.527, Q1 = 3.31, Median = 4.32, Q3 = 5.26, Maximum = 7.08; HER2-enriched (brown), n = 58, Minimum = 1.17, Q1 = 3.58, Median = 4.50, Q3 = 5.44, Maximum = 6.94; Normal-like (pink), n = 8, Minimum = 3.13, Q1 = 4.25, Median = 4.58, Q3 = 5.35, Maximum = 5.49; Basal-like (purple), n = 98, Minimum = 0.00, Q1 = 3.54, Median = 5.25, Q3 = 6.68, Maximum = 9.65. One-way ANOVA was used for statistical significance (F = 21.25, p<0.0001). FOXQ1 expression is expressed in RSEM (RNA-seq by expectation-Maximization) format. (B) Left, box plot of FOXQ1 RNAseq data in BC patients stratified by ER status. ER-positive (red), n = 601, Minimum = 0.00, Q1 = 2.82, Median = 4.12, Q3 = 5.05, Maximum = 8.13; ER-intermediate (orange), n = 2, Minimum = 4.24, Q1 = 4.24, Median = 4.50, Q3 = 4.50, Maximum = 4.50; ER-negative (blue), n = 179, Minimum = 0.00, Q1 = 3.58, Median = 4.68, Q3 = 6.26, Maximum = 9.65. One-way ANOVA was used for statistical significance (F = 24.64, p<0.0001). (B) Right, box plot of microarray data of FOXQ1 expression in BC patients stratified by ER status from the Van’t Veer et al48 study. ER-positive (red), n = 71, Minimum = −1.46, Q1 = −0.67, Median = −0.20, Q3 = 0.08, Maximum = 1.06; ER-negative (blue), n = 46, Minimum = −1.22, Q1 = −0.38, Median = 0.5, Q3 = 1.06, Maximum = 2.87. Welch’s t-test was used for statistical significance (***p<0.0001). Whiskers represent minimum and maximum values. The TCGA-BRCA95 and the Van’t Veer et al48 data were accessed using UCSC XENA cancer genome browser: https://xenabrowser.net/.