Figure 1. RADX inhibits RAD51 assembly on DNA.
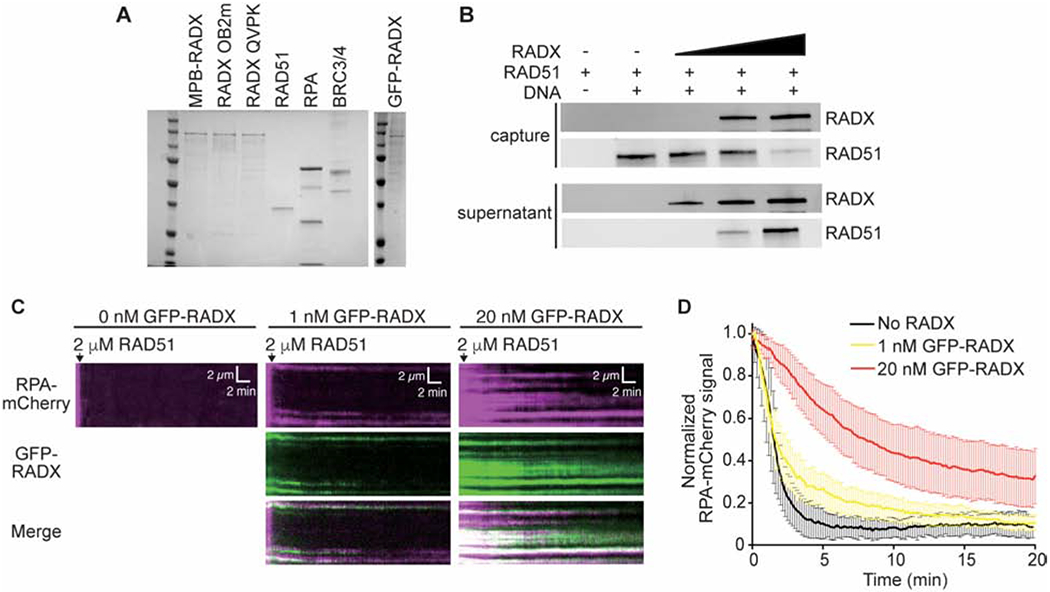
(A) Coomassie stained SDS-PAGE gel showing purified proteins used in this study. Each lane contains ~200 nM of purified protein. Actual amounts used is specified in each experiment. (B) Pull-down experiments showing MBP-RADX (0.5, 1, and 2nM) outcompetes RAD51 (4μM) for binding ssDNA. (C) Kymographs showing the assembly of RAD51 on ssDNA molecules coated with RPA-mCherry and 0 nM (left), 1 nM (middle), and 20 nM GFP-RADX (right). Assembly was initiated by injecting 2 μM RAD51 together with 2 mM ATP and 5 mM Ca2+ into sample chambers containing ssDNA molecules coated with RPA-mCherry and GFP-RADX while monitoring the loss of RPA-mCherry signal. (D) Graphs showing the normalized RPA-mCherry signal intensity integrated over entire ssDNA molecules during the assembly of the RAD51 filaments. Error bars represent 68% confidence intervals (n=30 ssDNA molecules).
