Figure 4. RADX interacts directly with RAD51 through residues in the RADX OB3 domain.
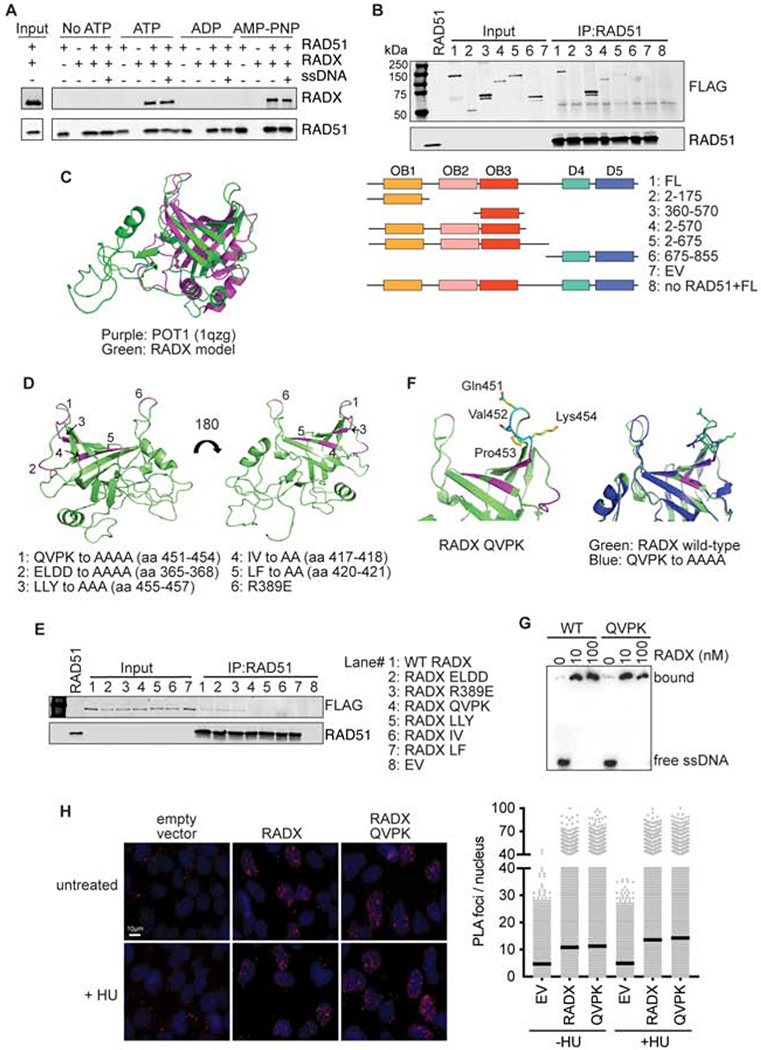
(A) Direct interaction of RADX and RAD51 was assessed using RAD51 antibody conjugated to Protein G beads to pull down purified RAD51 and RADX in the presence of the indicated nucleotide with and without ssDNA. Each binding reaction contains equimolar amounts of RADX and RAD51. After washing, the beads were boiled in SDS loading buffer and eluted protein detected by immunoblotting. (B) Interaction of purified RADX fragments with RAD51 in the presence of ATP. FL, full length (C) Homology model of RADX OB3 domain based on the OB-fold domain in POT1 (PDBID 1qzg). (D) Location of the surfaces of RADX OB3 tested for interactions with RADX. (E) Interaction of purified RADX OB3 mutants with RAD51 in the presence of ATP. (F) Location of the QVPK residues on the predicted loop of OB3 (left panel) and comparison of the models of wild-type and QVPK mutant OB3 domains (right panel). (G) Electrophoretic mobility shift assay of purified RADX and RADX QVPK mutant protein with poly (dT60) ssDNA. (H) Proximity ligation assay between Flag-RADX and EdU. Cells were labeled for 10 minutes with EdU and treated with hydroxyurea (HU) for two hours where indicated. An image and quantitative data from a representative experiment is shown. (EV, empty vector)
