Figure 1. Engineering PSC-derived epidermal organotypic rafts with a conditional FA pathway.
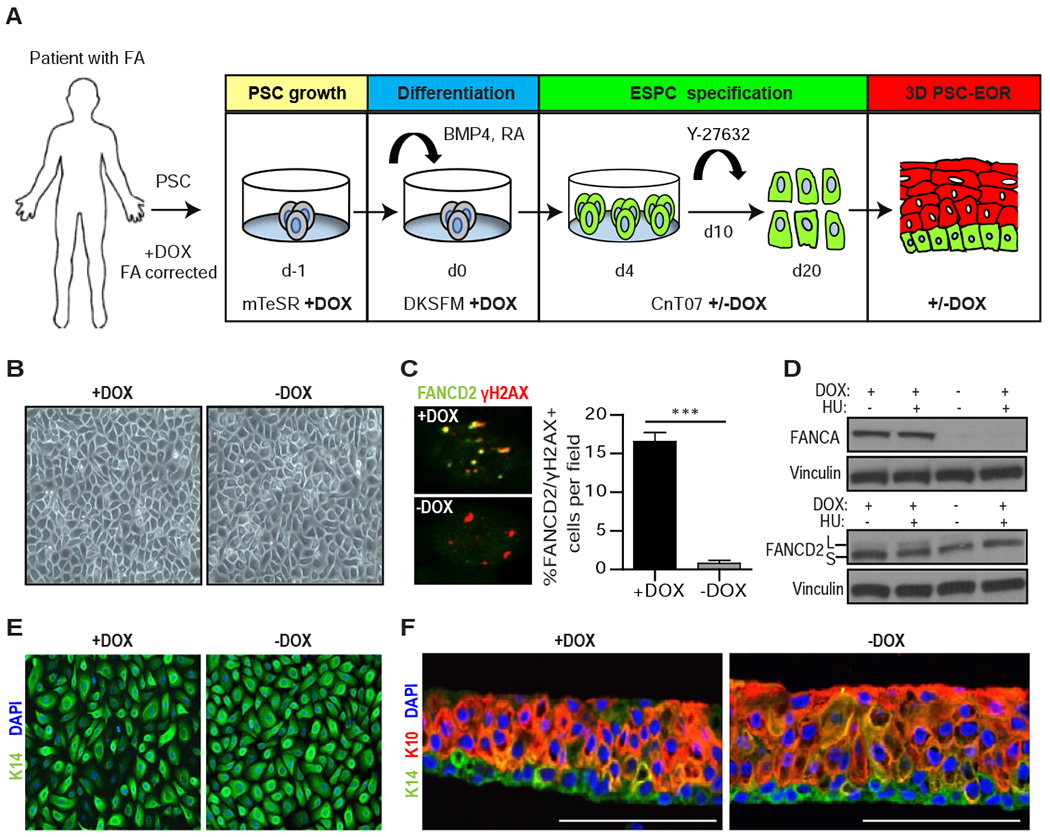
(A) Schematic of the 20-day protocol used for the directed differentiation of patient-derived cFA-PSCs into ESPCs and 3D engineered epidermis, with cells cultured in the presence of DOX starting on day −1. To generate FA pathway-deficient ESPCs, DOX was removed on day 4. mTeSR: embryonic/PSC medium; DKSFM: defined keratinocyte serum-free media; CnT07: progenitor-cell-targeted epidermal-keratinocyte media; Y-27632: ROCK kinase inhibitor. (B) Bright field images of cFA-ESPCs at passage 2 showing similar formation of cohesive sheets of cells; one representative example is shown. (C) Immunofluorescence (IF) analysis and proportion of FANCD2 and γH2AX+ cells in +/−DOX ESPCs treated with 300 nM MMC for 16 hours prior to fixation; ***P<0.0001, two-tailed Student’s t-test; n=3. Error bars represent SEM. (D) Western blot analysis of whole-cell lysates from +/−DOX ESPCs that were untreated or DNA-damaged with 2 mM hydroxyurea (HU) for 24 hours. FANCD2 monoubiquitination demonstrates the inducibility of the FA pathway. L and S: monoubiquitinated and unmodified forms of FANCD2, respectively. Vinculin, loading control; n=2, one representative example is shown. (E) IF analysis for the basal cell marker K14 and 4’6-diamidine-2-phenylindole (DAPI) in ESPCs at passage 2, demonstrating basal cell characteristics; n=3; Magnification, 40x. (F) PSC-EORs derived from +/−DOX ESPCs were processed for IF to image K14+ basal and K10+ suprabasal cells, and DAPI staining. Scale bar, 100 μm. See also Figure S2.
