FIGURE 3.
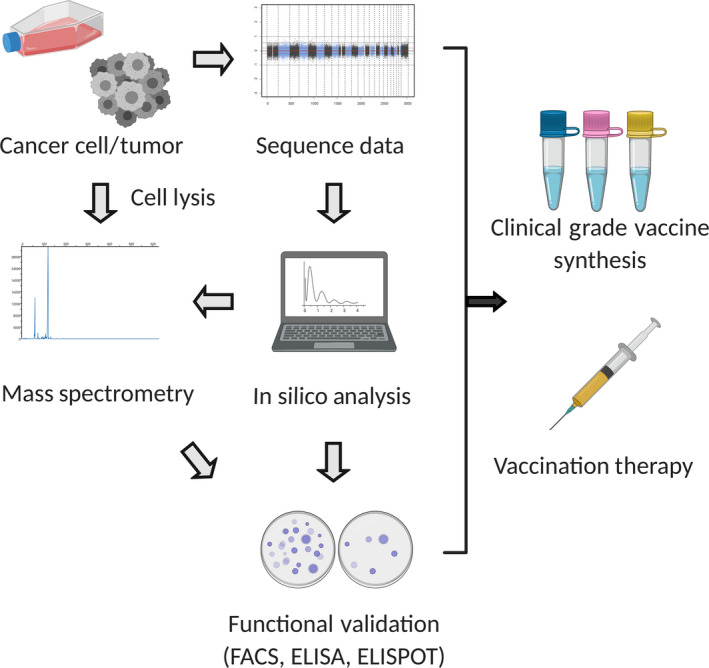
Pipeline for neoantigen identification. The sequence data (whole exome and RNA sequencing data) identifies neoantigen candidates by single nucleotide variants or insertions/deletions. Gene expression data allows narrowing down of expressed candidates. In silico analysis is then carried out with prediction software. In most cases, high‐affinity (IC50 < 500 nmol/L) candidates are selected. Mass spectrometry is an approach to validate candidates based on MHC bound peptides from tumor/cell lysis extraction. Functional analysis including FACS, ELISA, and ELISPOT further validate neoantigen candidates. After the integration of information, peptides/DNA/RNA vaccines are synthesized
