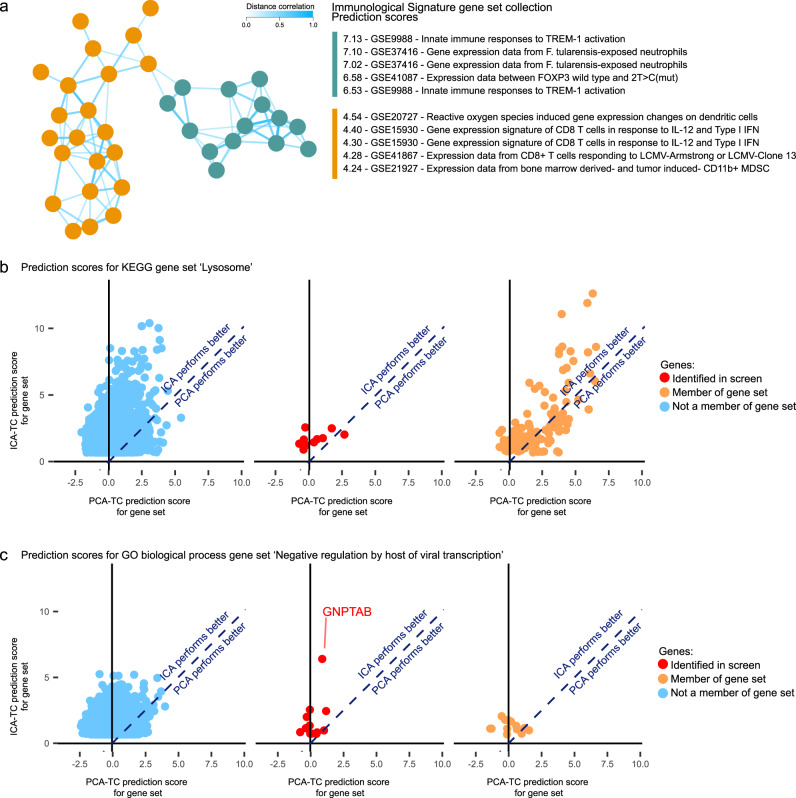
Fig. 8. GENETICA predictions for other CRISPR-based screens.
a Co-functionality network generated with 111 genes prioritized in a CRISPR screen investigating the inflammatory response to bacterial lipopolysaccharide. Two clusters were formed that showed enrichment for predicted involvement in an inflammatory response (right cluster) and oxidative stress response in dendritic cells (left cluster). Predictions in this co-functionality network were based on the Immunological Signatures gene set collection. Only genes with a co-functionality above a threshold of R > 0.5 are shown in the network. b Scatterplots of ICA-TC-based (y-axis) and PCA-TC-based (x-axis) prediction scores of all genes for the KEGG lysosome gene set. The orange- and blue-colored dots designate the membership of genes to the respective KEGG lysosome gene set, and red dots designate genes identified in the CRISPR screen mentioned in the text. c Scatterplots of ICA-TC-based (y-axis) and PCA-TC-based (x-axis) prediction scores of all genes for the GO Biological Process gene set Negative regulation of viral transcription by the host. The orange and blue color dots designate the membership of genes to the respective GO Biological Process gene set, and red dots designate genes identified in the CRISPR screen mentioned in the text; the dot corresponding to the GNPTAB gene is highlighted.