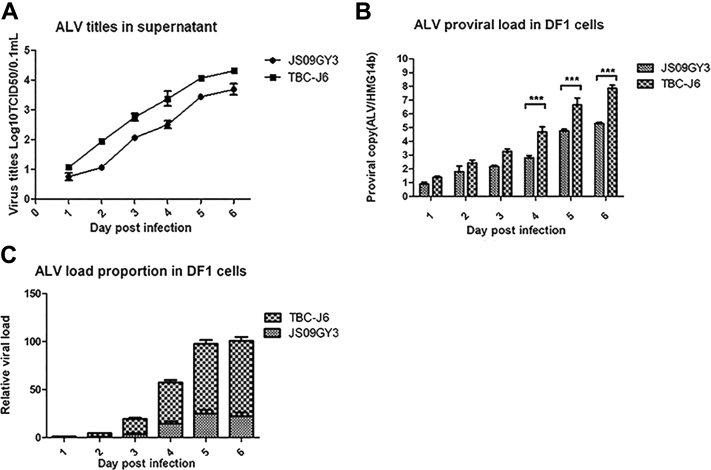
Figure 4.
Comparison of proliferation and competitive advantages of different strains. (A) Supernatants of different infected cells were harvested at 1 to 6 d after infection and then titered for 50% tissue culture infective dose (TCID50) using the Spearman–Kärber method. (B) DNA of the infected cells was extracted at 1 to 6 d after infection. The proviral genome load in DF-1 cells was detected by real-time quantitative PCR. HMG14b is used as a reference. (C) RNA of the cells coinfected with 2 viruses was extracted at 1 to 6 d after infection. Quantitative reverse transcription PCR was performed to test the expression of the 2 viruses at different intervals, with 18s RNA used as a reference.