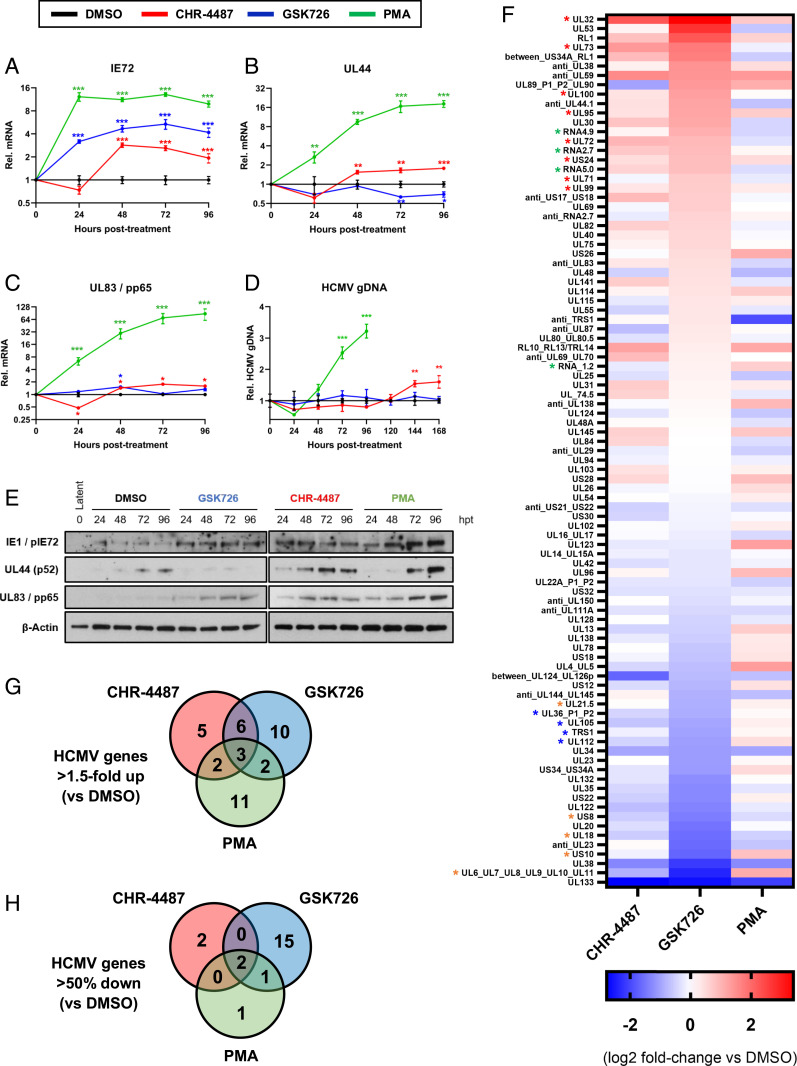
Fig. 3.
BET inhibitors dysregulate HCMV transcription from latency and restrict viral DNA replication. (A–E) CD14+ monocytes isolated from apheresis cones infected with HCMV TB40e wild type for 5 d were treated with DMSO (black lines), HDACi (CHR-4487 30 nM, red lines), I-BET (GSK726 30 nM, blue lines) and PMA (20 nM, green lines). RNA, DNA, and protein were isolated from cells every 24 h for 4 d. RT-qPCR was then performed for virus (A) IE72, (B) early UL44, and (C) late UL83/pp65 transcripts relative to host GAPDH with (D) HCMV genome copy number determined by qPCR of UL44 promoter relative to host GAPDH promoter and (E) immunoblotting carried out for virus targets relative to β-actin (images representative of n = 3). *P < 0.05, **P < 0.01, ***P < 0.001; mean ± SEM, n = 3. (F–H) RNA-seq of flow cytometry isolated YFP-positive HCMV-TB40e-IE86-eYFP–infected CD14+ monocytes after 72 h treatment with compounds (F). Data are presented as a heat map of means of HCMV transcripts identified from biological duplicates, with data ranked by I-BET (GSK726) fold-change relative to DMSO control. HCMV gene groups indicated by associated asterisk color (red, structural protein; green, long noncoding RNA; orange, immunomodulatory protein; blue, DNA replication protein). Venn diagrams show HCMV transcript number from RNA-seq data either (G) up-regulated (>1.5-fold) or (H) down-regulated (>50%) relative to DMSO control.