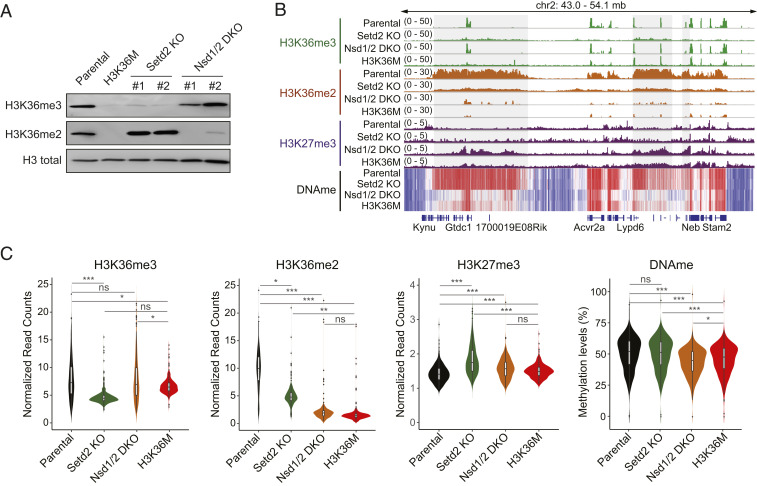
Fig. 1.
Generation and epigenome profiling of Nsd1/2 DKO and Setd2 KO 10T cells. (A) Western blotting of H3K36me3 and H3K36me2 levels in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO 10T cells. Total H3 is used as loading control. For Nsd1/2 DKO and Setd2 KO, clone 1 is used for all epigenome profiling analyses. (B) Integrative Genomics Viewer snapshot showing the chromatin landscape of H3K36me3, H3K36me2, H3K27me3, and DNA methylation at a representative genomic region in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO 10T cells. (C) Violin plots showing the global levels of H3K36me3, H3K36me2, H3K27me3 (counts per million ChIP-seq reads normalized to reference chromatin spike-in control), and DNA methylation (average methylation level) in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO cells. Bin size, 10 Mb. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.