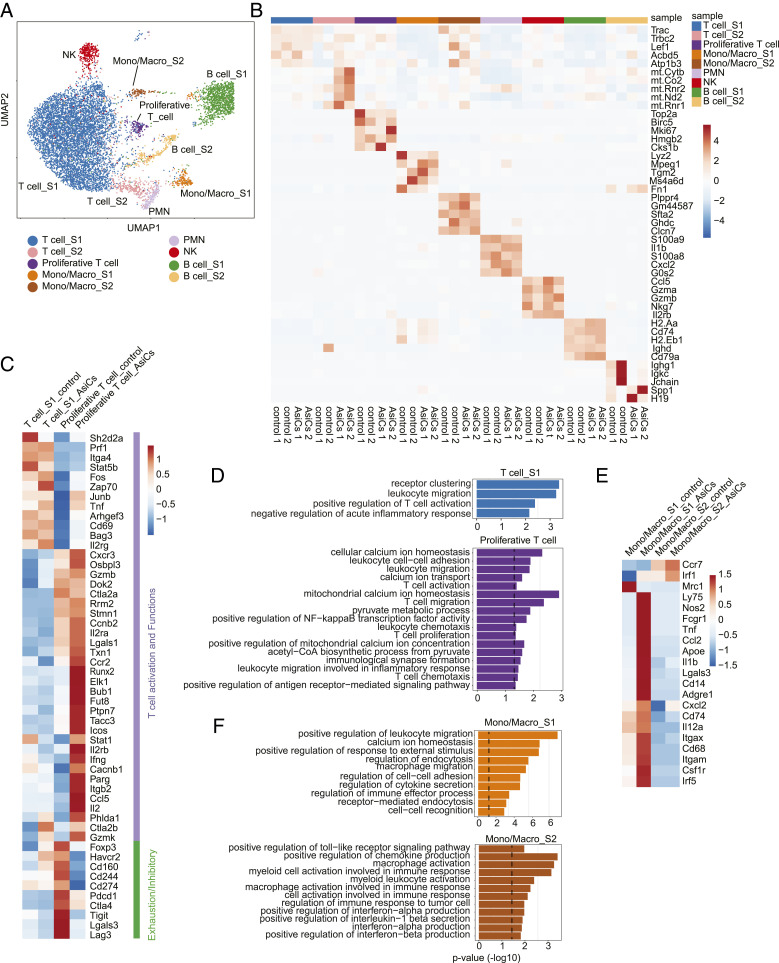
Fig. 5.
Functional improvement of tumor-infiltrating immune cells after treatment with immune-modulating EpCAM-AsiC as analyzed by scRNA-seq. CD45+ cells from 4T1E tumors in mice treated with EpCAM aptamer or EpCAM-AsiC targeting Upf2, Cd47, Mcl1, and Parp1 were pooled (n = 2) for scRNA-seq analysis. (A) Uniform manifold approximation and projection (UMAP) plot of CD45+ immune cells in all four samples. Each dot represents a cell that was colored by inferred cluster identity. (B) Heatmap showing the z-score–normalized expression of canonical cell-type DEGs across different clusters. Each column represents a biological sample. (C and D) Heatmap showing expression of genes involved in T cell activation, effector function, memory and exhaustion in T cell clusters, averaged per cluster and z-score standardized across clusters (C) and GO enrichment for DEGs up-regulated in EpCAM-AsiC-treated T cells (D). (E and F) Heatmap showing expression of genes involved in monocyte/macrophage phenotype and function in monocyte/macrophage clusters, averaged per cluster and z-score standardized across clusters (E) and GO enrichment for DEGs up-regulated in EpCAM-AsiC–treated monocytes/macrophages (F). Dashed lines in D and F indicate P = 0.05 (one-sided Fisher’s exact test).