Figure 8.
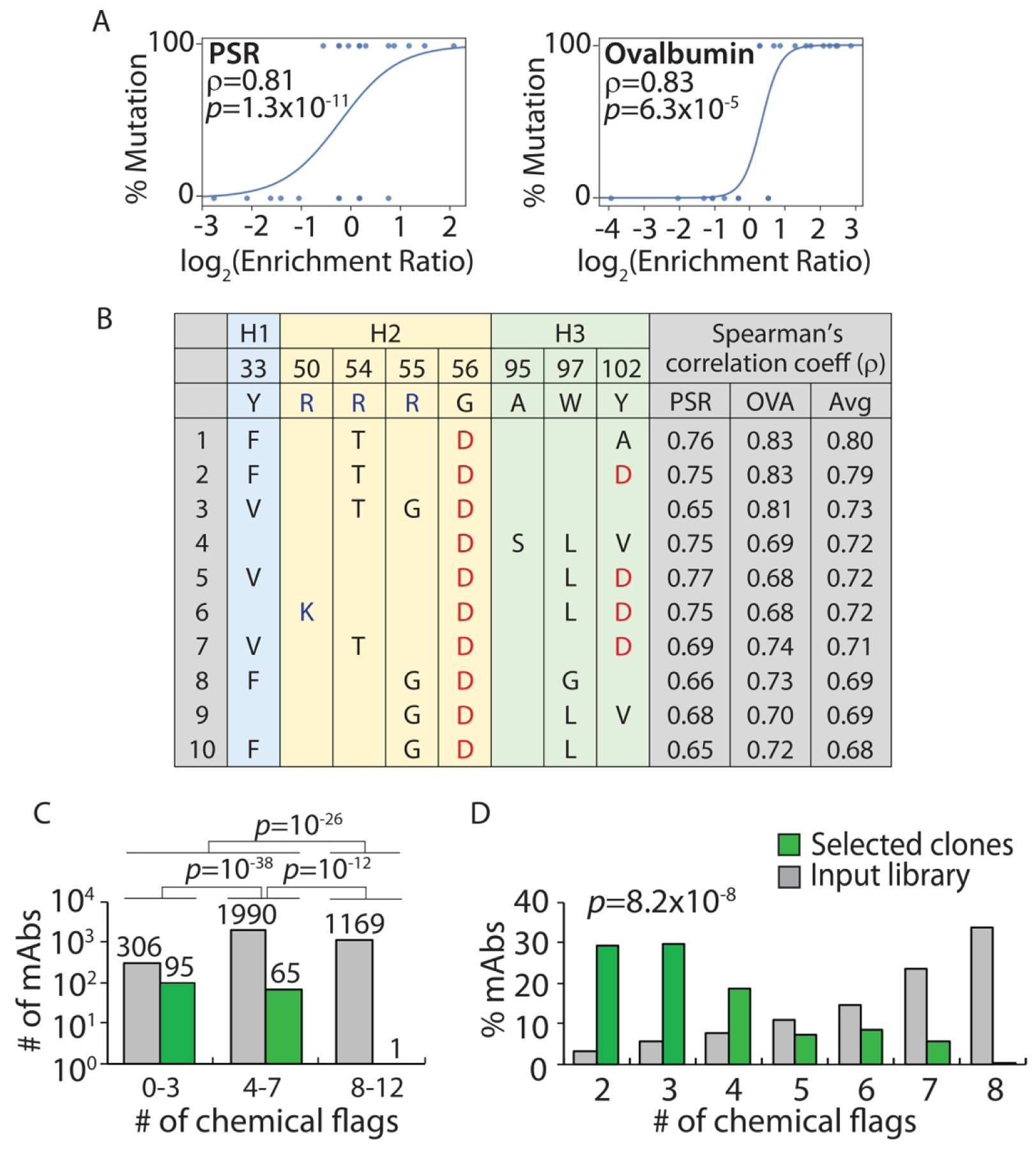
Design of Fab sub-libraries of emibetuzumab guided by the combined chemical rules and evaluation of selected mutants with improved antibody specificity. The VH domain of emibetuzumab was mutated at eight solvent-exposed sites (Y33, R50, R54, R55, G56, A95, W97 and Y102) in the three heavy chain CDRs that were flagged by the maximum chemical rules. The mutations sampled the wild-type residue as well as five mutations that are predicted to reduce the number of chemical flags. The libraries were constructed as single-chain Fab fragments (scFabs) on yeast, sorted for non-binding to two polyspecificity reagents [PSR and ovalbumin (OVA)], and evaluated via deep sequencing. (A) Enrichment ratios for antibody variants with a set of four mutations (F33, T54, D56 and A102 in VH) relative to antibody variants with wild-type residues at the same positions (Y33, R54, G56 and Y102 in VH) for two different polyspecificity reagents. The curves (logistic regressions) are guides to the eye. (B) Top ten sets of four mutation combinations that are most strongly correlated with reduced binding to polyspecificity reagents and increased specificity. (C, D) The (C) number and (D) percentage of mAb variants selected with high specificity as a function of the number of chemical flags relative to the corresponding values for the input library. In (A), the mAbs included in the wild-type or mutant groups are only required to have wild-type or mutant residues at the four evaluated sites and can have either wild-type or mutant residues at the other four sites. Moreover, the p-values are for the Spearman’s correlation coefficients (ρ). In (C), the p-values for the comparisons of the number of mAbs were calculated using a 2×2 contingency table (Fisher’s exact test). In (D), the p-value for comparing the distributions of mAbs was calculated using paired sample t-test (two tailed).
