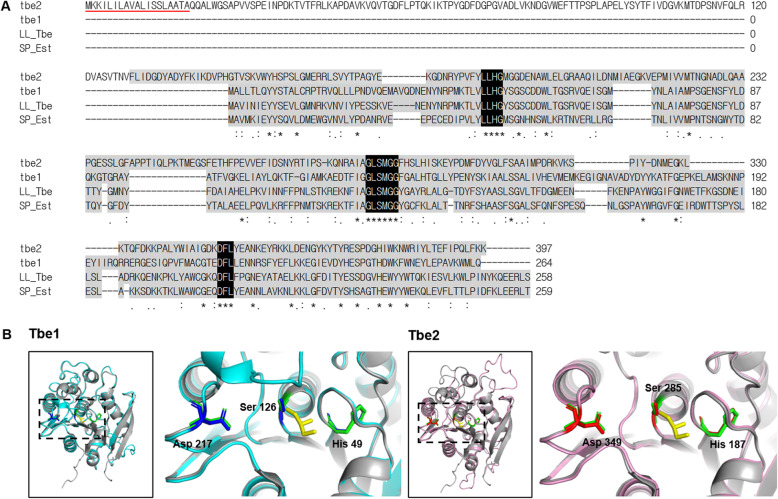
Fig. 3.
Amino acid sequences and 3-dimensional structures of Tbe1 and Tbe2. a Amino acid sequences and multiple amino acid sequence alignment of Tbe1, Tbe2, TB esterase of L. lactis (LL_Tbe; UniProtKB Q9L9X0), and esterase of Streptococcus pneumoniae (SP_Est; UniProtKB A0A0H2UNZ8). Alpha/beta hydrolase conserved domains and significant consensus sequences are highlighted in grey and black, respectively. Signal peptide is underlined in red. b Partial structure of Tbe1 and Tbe2 modelled by SWISS-MODEL. The models are based on the structure of the TB esterase of Streptococcus pneumoniae from the RSC Protein Data Bank (2UZ0) as the template. Tbe1, Tbe2, and S. pneumoniae TB esterase are shown in cyan, pink, and grey, respectively, with the glycerol residue of TB in yellow. Inside the dotted box is the structure containing the serine-histidine-aspartate (SHD) catalytic triad, as magnified on the right. Each SHD triad of Tbe1, Tbe2 and S. pneumoniae tributyrin esterase are shown in blue, red, and green, respectively