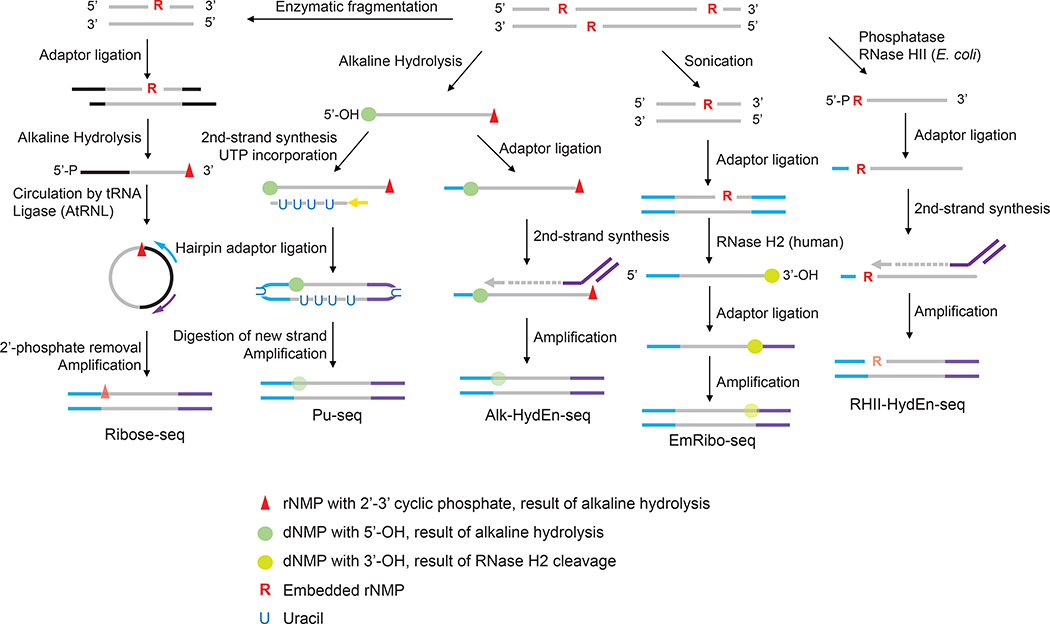
Figure 1.
The current iterations of ribonucleotide mapping technologies. These strategies can be classified by the methods used to incise at embedded ribonucleotides. Ribose-seq, Pu-seq and Alk-HydEn-seq use alkaline hydrolysis which catalyzes hydrolysis on the 3’ side of the rNMP, resulting in 2’,3’-cyclic phosphate and a 5’-hydroxyl DNA ends. EmRibo-seq uses recombinant human RNase H2 and RHII-HydEn-seq uses E. coli type II RNase H (RNase HII), both of which cleave on the 5’ side of the rNMP, resulting in 5’-phosphate and 3’-OH ends. These technologies also differ in the location of ribonucleotide with respect to the sequencing read. Ribose-seq maps the rNMP site to the first position of the mapped read but on the opposite strand. Pu-seq and Alk-HydEn-seq identify the rNMP as located one nucleotide upstream of the mapped the read. In EmRibo-seq, the rNMP is similarly positioned one base upstream of the mapped read but on the opposite strand. For RHI-HydEn-seq, the rNMP is the first nucleotide of the mapped read. The symbols denoting the DNA/RNA ends resulting from alkaline hydrolysis or RNase cleavage are kept throughout the steps to help track their locations in the sequencing reads. It is worth noting that after the DNA amplification step during the library preparation, these symbols no longer represent the original form of the nucleotides but rather their base identities. Their opacity to emphasize this difference.