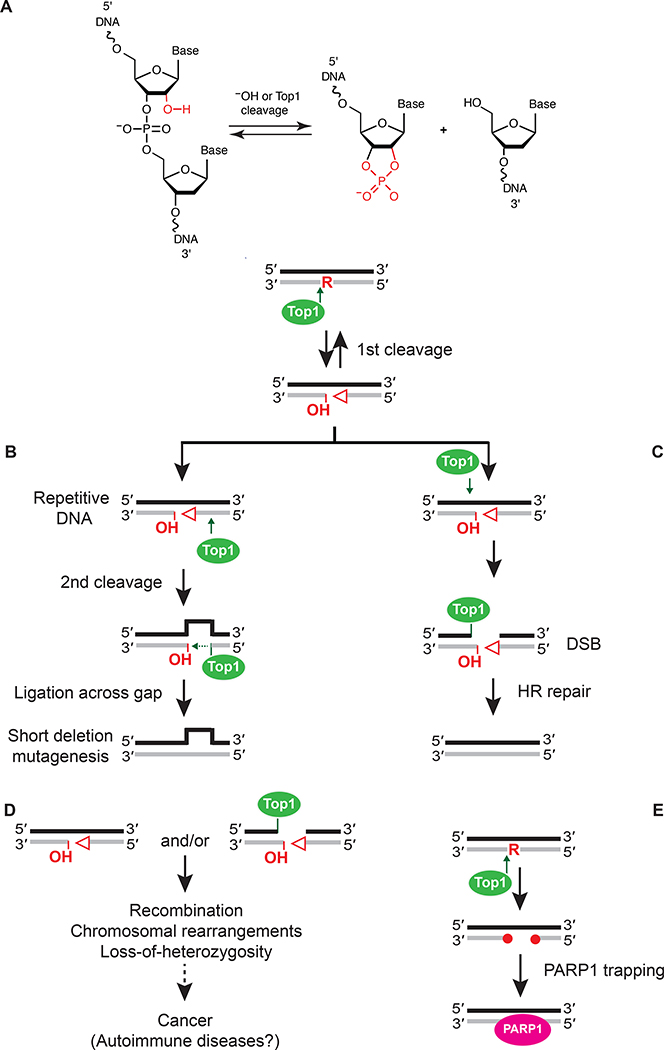
Figure 3.
The negative physiological consequences associated with processing of ribonucleotides in DNA. (A) A single ribonucleotide in DNA can lead to formation of a single strand break (SSB) containing unligatable 5’-OH and 2’,3’-cyclic phosphate DNA ends (open triangle). This can occur following spontaneous hydrolysis or Top1 cleavage. (B) Short deletions in repeat DNA sequences are likely generated by two consecutive Top1 cleavage events, the first at the site of a ribonucleotide and the second 5’ to this initial cleavage site. This allows for strand realignment and Top1-mediated ligation across the short gap, resulting in loss of a repeat DNA sequence. (C) A DNA DSB can be directly generated following two Top1 cleavage events. If the initial Top1 cleavage event at an unrepaired ribonucleotide occurs in close proximity to a Top1 cleavage event on the opposite DNA strand, this generates a DSB that is repaired by Rad51- and Rad52-mediated HR. (D) Processing of genomic ribonucleotides results in SSBs and DSBs with unligatable DNA ends that promote various types of genome instability and may contribute to diseases such as cancer and autoimmune disorders. (E) Aberrant DNA ends produced following Top1-cleavage at unrepaired genomic ribonucleotides are recognized and bound by PARP1 to initiate DNA repair.