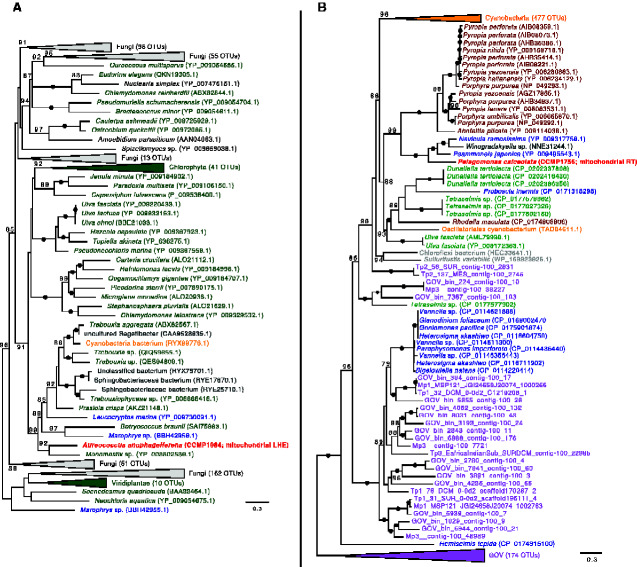
Fig. 4.
Maximum-likelihood phylogeny of (A) the group I intron LAGLIDADG homing endonuclease (LHE) encoded in coxI in Aureococcus anophagefferens and (B) the group II intron encoded reverse transcriptase (RT) in the rnl gene in Pelagomonas calceolata. Pelagophyte sequences are bolded and colored red. Cyanobacterial homologs are indicated in orange, whereas other bacterial/archaeal homologs are indicated in gray. Viral homologs are indicated in pink (nr database) or purple (Tara Global Ocean Viromes [GOV] dataset). Other colors represent eukaryotic homologs. Numbers in brackets indicate the number of homologs collapsed at a given node. Numbers on branches indicate standard bootstrap support; only values higher than 70 are shown and black circles indicate maximal support for a particular node. The tree is midpoint rooted and was inferred using 159 (LHE) and 587 (RT) unambiguously aligned sites. The scale bar shows the number of inferred amino acid substitutions per site.