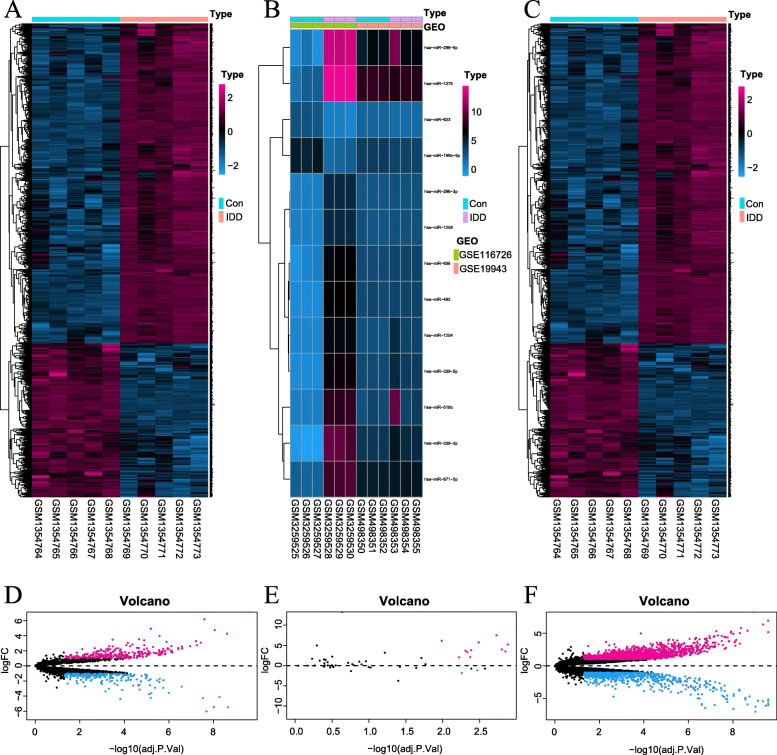
Fig. 3.
Difference analysis. a-c The differentially expressed heat-maps of lncRNA, microRNA, and mRNA. The lncRNA heat-map of GSE56081 dataset (a); The merged microRNA heat-maps of GSE116726 and GSE19943 datasets (b); The mRNA heat-map of GSE56081 dataset. d-f The volcano plot (c). The lncRNA dataset of GSE56081 (d); The merged microRNA datasets of GSE116726 and GSE19943 (e); The mRNA dataset of GSE56081 (f). The screening condition: absolute values of log (fold change) > 1.0 and adjusted p-Value < 0.05. In the plots, the rose red and blue dots represent high and low RNA expressions with statistical significance between the IDD NPs and the normal NPs, respectively. And, the black dots represent no statistical significance between them