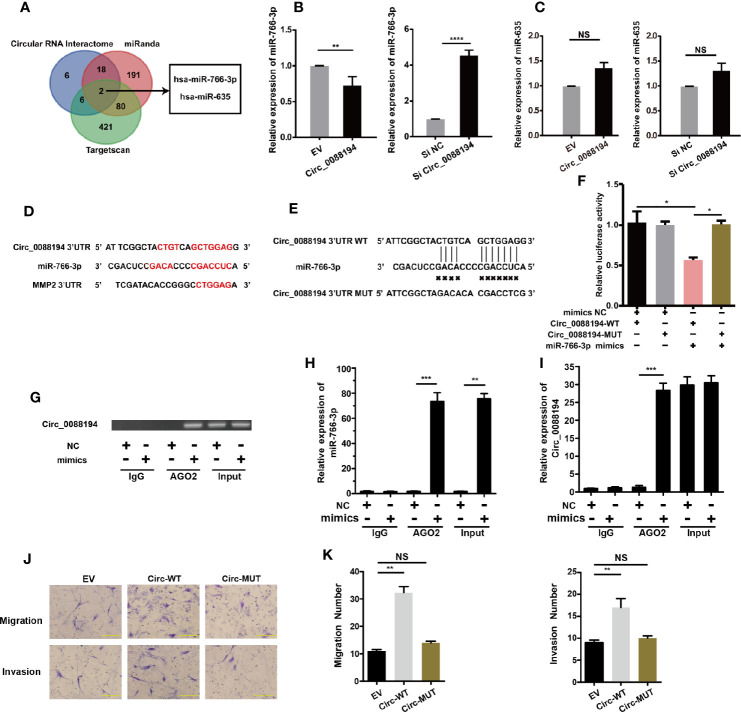
Figure 4.
Circ_0088194 act as sponge of miR-766-3p in RA-FLSs. (A) Schematic illustration showing the identification of two miRNAs, hsa-miR-766-3p and hsa-miR-635, predicted from three available databases (miRanda, circular RNA interactome, and Targetscan). (B, C) QRT-PCR determination of the expression of miR-766-3p (B) and miR-635 (C) in RA-FLSs. The cells were transfected with adenovirus expressing Circ_0088194 or Circ_0088194#1 siRNA. (D) The miR-766-3p MREs in the sequences of Circ_0088194 and MMP2 as identified using bioinformatic analysis. (E) Wild-type (WT) or mutant (MUT) miR-766-3p target sequences of Circ_0088194. (F) The miR-766-3p mimics reduced the luciferase activities of WT not MUT Circ_0088194 reporter genes, as assessed using dual-luciferase reporter assays. (G) RA-FLSs were transfected with the miR-766-3p mimics or the negative control. Anti-AGO2 RIP was used to investigate AGO2 binding to Circ_0088194 and miR-766-3p; IgG was used as a negative control. (H, I) The expression levels of Circ_0088194 and miR-766-3p, as detected using qRT-PCR. (J, K) The migratory and invasive activities of RA-FLSs were evaluated by Transwell migration and Matrigel invasion assays. RA-FLSs were transfected with the adenovirus encoding Circ_0088194 or the adenovirus encoding Circ_0088194 with mutated miR-766-3p binding sites. Scale bar, 500 µm. Data are shown as mean ± SD. NS, not significant. *indicates p < 0.05, **indicates p < 0.01, ***indicates p < 0.001, and ****indicates p < 0.0001.