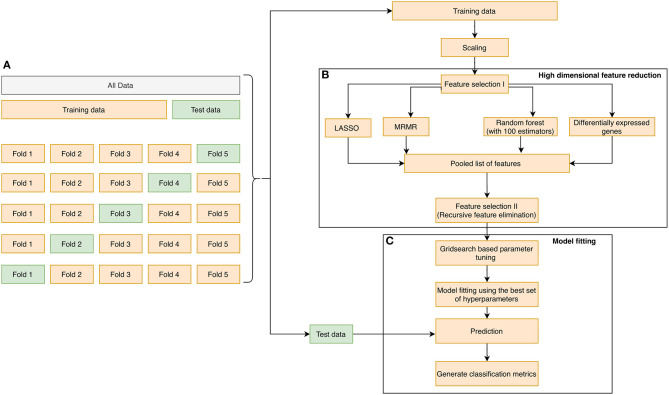
Figure 1.
The overall methodology design for biomarker discovery from the derivation dataset GSE66099 containing 228 samples is illustrated in this figure. (A) The initial data is aggregated, normalized, corrected for batch normalization, and separated into even chunks using k-fold cross-validation (CV). In our pipeline, we used k = 5. (B) The training chunks of the CV are used for model development; the data analysis pipeline follows the Complete Cross-Validation (CCC) approach defined by Alder et al. (42). In addition to DEG, we apply three other variable selection methods to generate a pool of candidate genes. We then apply a wrapper method, namely the RFE to arrive on the most predictive genes. (C) The genes selected by the RFE method are then used to develop a predictive model. The model is then evaluated on the test fold of the CV. This process is repeated for the remaining training and test folds. Finally, the entire 5-fold CV is repeated 10 times to generate a total of 50 iterations, and the top predictors from (B) are saved and analyzed to generate a normalization score, which is a measure of how often a gene appears as a top predictor across each of the 50 iterations.