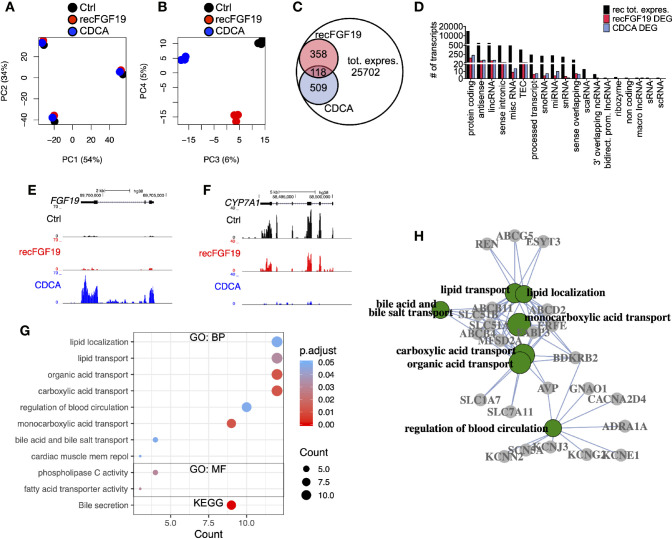
Figure 4.
Differentially expressed genes in primary human hepatocytes upon treatment with chenodeoxycholic acid (CDCA) or recombinant FGF19. Total RNA from primary human hepatocytes treated with recombinant FGF19, CDCA, or vehicle control was sequenced (n = 3). (A, B) Principal component analysis (PCA) showed that 88% of the variation in the samples could be explained by donor differences (PC1-2), while 11% of the variation could be explained by the treatment (PC3-4). (C, D) A limited number of transcripts were differentially expressed upon treatment. Venn diagram (C) and bar plot (D) displaying all expressed transcripts (black) and differentially expressed ones after recombinant FGF19 treatment (red) or CDCA treatment (blue). (E, F) Representative UCSC Genome Browser tracks of normalized FGF19 and CYP7A1 expression after recombinant FGF19 or CDCA treatment. The tracks from top to bottom show the scale in the human genome, the location in the human genome, the gene including exons (black boxes) and introns (arrows), and the RNA-seq signal from each treatment. (G) Unique chenodeoxycholic acid (CDCA)-DE transcripts were used for gene ontology (GO) biological processes (BP), molecular function (MF), and KEGG pathway analysis. Displayed are all significant ontologies/pathways in each ontology. The size of the bubble indicates number of genes in each category and the color represents the significance. (H) GO-term interaction network of the 7 most significant GO-BP terms in (G). GO terms are in green circles, and gene names are in grey circles. Unique DE transcripts after recombinant FGF19 gave no significant enrichment in GO or KEGG pathway-related terms.