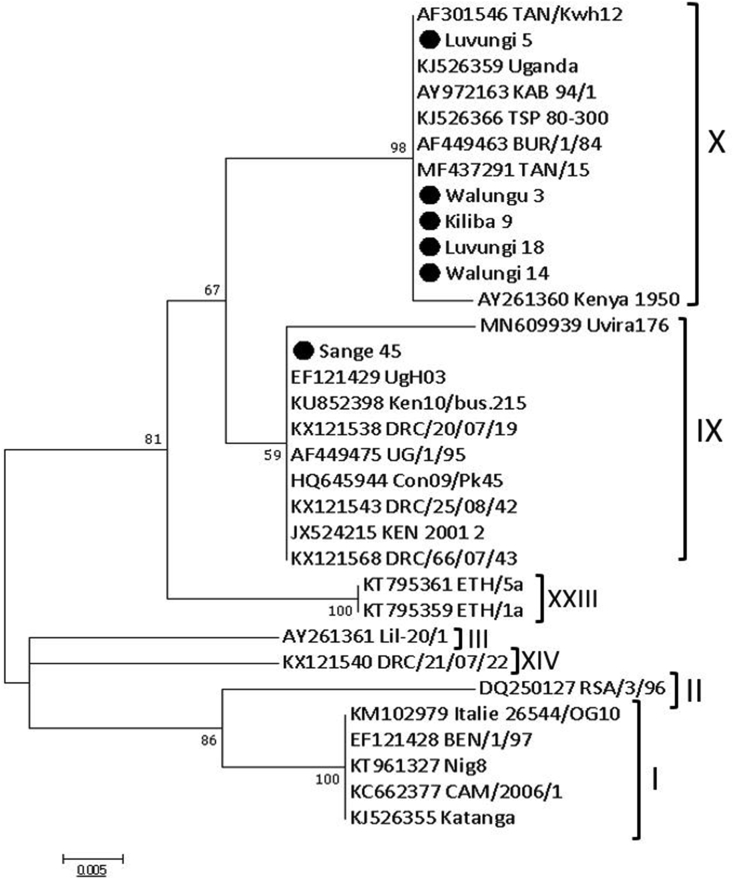
Figure 4.
Phylogenetic relationships of p72 genotypes. The evolutionary history was inferred by the maximum likelihood method based on the Kimura 2-parameter model (Kumar et al., 2016). The phylogeny was inferred following 1,000 bootstrap replications, and the node values show percentage bootstrap support. Strain names such as Kiliba 9, Luvungi 5, Luvungi 18 and Sange 45 are viruses from Uvira districts while Walungu 3 and Walungu 14 are strains from Walungu district. The scale bar indicates nucleotide substitutions per site. Dark-spot circles (plain circle •) represent sequences from this study.