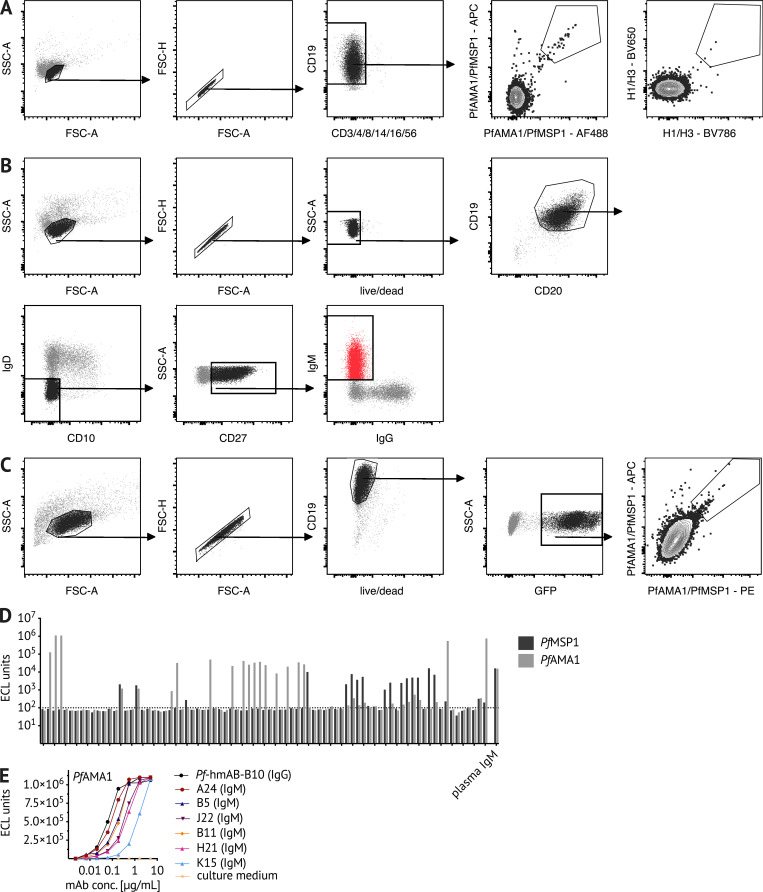
Figure S2.
Sorting strategies for BCR sequencing and immortalization of Pf-specific IgM B cells. (A and B) Representative flow cytometry plots of Malian PBMCs after B cell enrichment, showing gating on (A) live CD3−, CD4−, CD8−, CD14−, CD16−, CD56−, CD19+ Pf+ or HA+ B cells that were single-cell sorted for BCR sequencing, and (B) live CD19+ CD20+ IgD− CD10− CD27+ IgM+ B cells that were bulk sorted for transduction with the pLZRS-IRES-GFP retroviral vector expressing Bcl-6 and Bcl-xL (a gift from Lynda Chin, Addgene plasmid #21961; Kim et al., 2006). (C) 5 d after transduction, B cells were stained with Pf probes. Representative flow cytometry plots of transduced B cells showing gating on live CD19+ GFP+ B cells expressing GFP. PfAMA1 or PfMSP1 probe-binding cells were single-cell sorted into 384-well plates for continuous in vitro culture and BCR sequencing. (D) When tested for binding to PfMSP1/PfAMA1 using the MSD assay, 46.9% of culture supernatants (38 out of 81) bound the B cell probe target protein above the background threshold of 100 electrochemiluminescence (ECL) units. Data are from at least two independent experiments. (E) For six PfAMA1-binding clones, the MSD assay was used to quantify the binding of culture supernatant-derived IgM. PfAMA1-binding data from at least two independent experiments are displayed for six IgM clones (A24, B5, J22, B11, H21, and K15) and the IgG mAb clone B10. FSC, forward scatter; SSC, side scatter.