Figure 1 |. Example Allele-Differential Phenomenon in Common MPRA Approaches, and Analysis of MPRA Data.
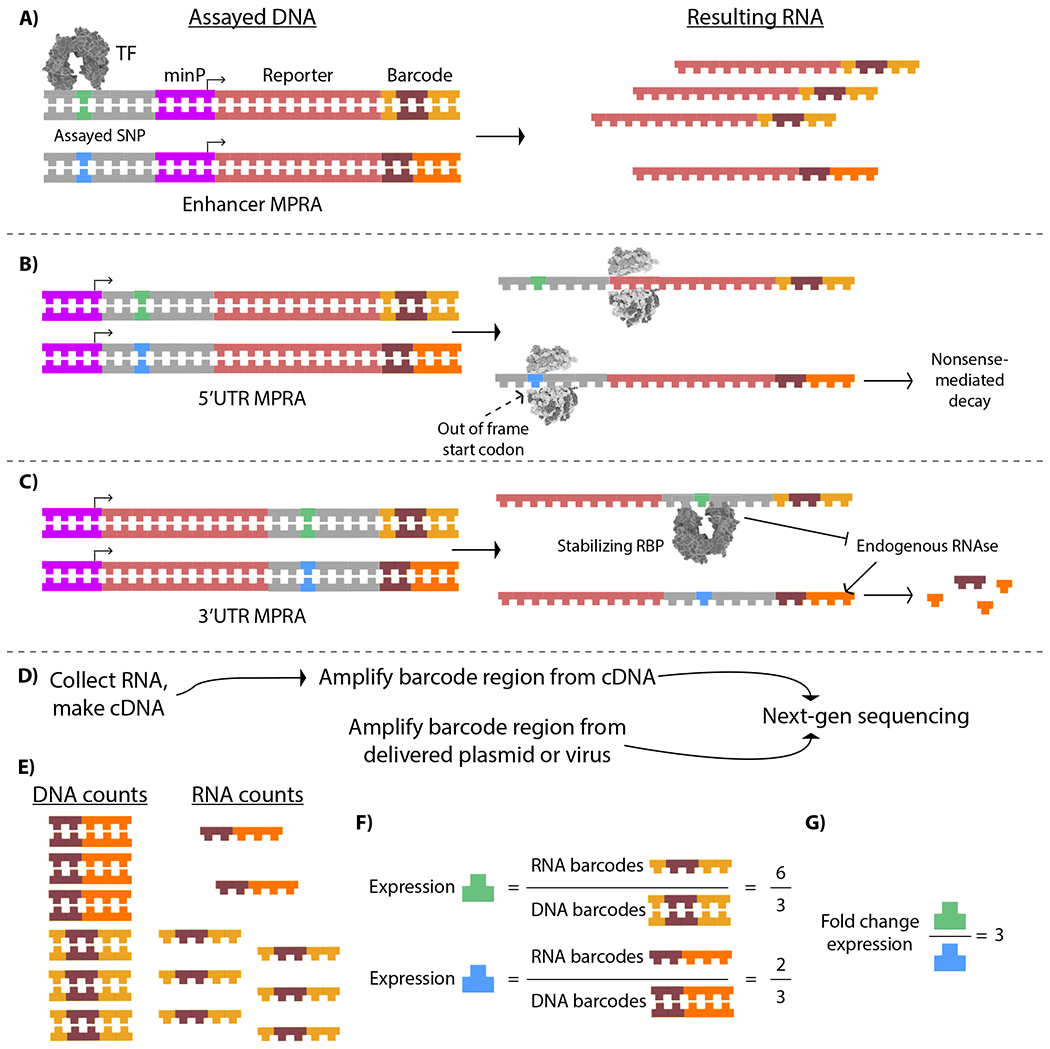
A) In a transcriptional-regulatory assay, a putative regulatory SNP may create, ablate, strengthen, or weaken a TF binding site. As a result, one allele drives more transcription (detected via its 3’UTR barcodes) per encoding DNA than the other allele. B) In a 5’UTR assay, a functional SNP may sequence features controlling translation initiation. For example, a variant allele may introduce an upstream start codon out of frame with the reporter gene, resulting in nonsense mediated decay, and thus, decreased detection of the barcodes paired to that UTR allele. C) In a 3’UTR assay, a variant may alter an RBP binding site; in this example, an RBP site specific to one allele increases the stability of the reporter transcript, and thus of the barcode paired to it. D) After transfection/transduction, RNA is collected from specimens and prepared along with DNA (often the delivered DNA, though sometimes this is recovered from the specimens as well) to generate sequencing libraries to quantify expression of the delivered elements in the RNA, compared to starting abundance in the DNA. E) Example read counts, presented visually, for the DNA and RNA barcode counts of one barcode paired to each allele. F) MPRA analysis centers on taking the ratio of RNA/DNA counts (or counts per million), represented by the sequence fragments at top left, as a measure of expression—i.e., approximating the number of transcripts generated per encoding DNA. These can be compared relative to the expression of all elements to find the strongest features (e.g., strongest enhancers and repressors, or most stabilizing and destabilizing UTR elements), or G) compared on a variant-wise basis to determine significant allelic regulatory effects.
