FIGURE 5.
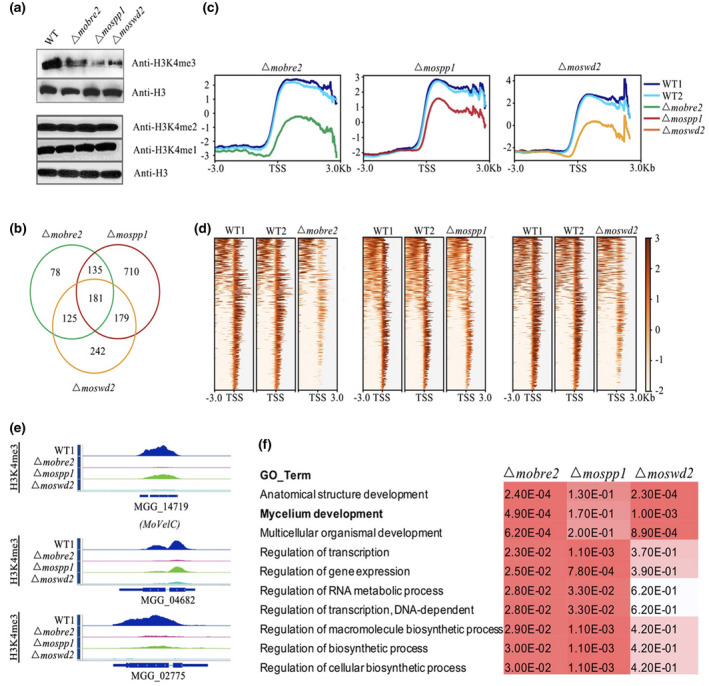
The MoBre2, MoSwd2, and MoSpp1 deletion mutants display significantly decreased H3K4me3 profiles in transcription start site (TSS) overlapped target regions. (a) Western blot analysis of histone H3K4me3, H3K4me2, and H3K4me1 modifications in the wild‐type (WT) P131 and Δmobre2, Δmospp1, and Δmoswd2 strains. (b) The number in the Venn diagram shows the H3K4me3 decreased peaks in the deletion mutants compared with the WT. (c) The H3K4me3‐ChIP‐seq distribution of enriched peaks around the TSSs of the overlapped genes in MoBre2, MoSwd2, and MoSpp1 deletion mutants. (d) The heatmaps show the distribution of H3K4me3‐ChIP‐seq peak signal density in different samples. (e) The read coverage shows that the peaks in these three deletion mutants were significantly decreased compared with the WT strain in some sample genes. (f) Gene ontology (GO) term comparisons of the down‐regulated genes of deletion mutants, especially in spore germination and pathogenesis in the Δmobre2, Δmospp1, and Δmoswd2 mutants
