Abstract
Soybean (Glycine max) is one of the most important cultivated plants worldwide as a source of protein‐rich foods and animal feeds. Anthracnose, caused by different lineages of the hemibiotrophic fungus Colletotrichum, is one of the main limiting factors to soybean production. Losses due to anthracnose have been neglected, but their impact may threaten up to 50% of the grain production.
Taxonomy
While C. truncatum is considered the main species associated with soybean anthracnose, recently other species have been reported as pathogenic on this host. Until now, it has not been clear whether the association of new Colletotrichum species with the disease is related to emerging species or whether it is due to the undergoing changes in the taxonomy of the genus.
Disease symptoms
Typical anthracnose symptoms are pre‐ and postemergence damping‐off; dark, depressed, and irregular spots on cotyledons, stems, petioles, and pods; and necrotic laminar veins on leaves that can result in premature defoliation. Symptoms may evolve to pod rot, immature opening of pods, and premature germination of grains.
Challenges
As accurate species identification of the causal agent is decisive for disease control and prevention, in this work we review the taxonomic designation of Colletotrichum isolated from soybean to understand which lineages are pathogenic on this host. We also present a comprehensive literature review of soybean anthracnose, focusing on distribution, symptomatology, epidemiology, disease management, identification, and diagnosis. We consider the knowledge emerging from population studies and comparative genomics of Colletotrichum spp. associated with soybean providing future perspectives in the identification of molecular factors involved in the pathogenicity process.
Useful website
Updates on Colletotrichum can be found at http://www.colletotrichum.org/.
All available Colletotrichum genomes on GenBank can be viewed at http://www.colletotrichum.org/genomics/.
Keywords: Colletotrichum truncatum, emerging diseases, fungal pathogens, Glomerella, Glycine max
A revision of soybean anthracnose revealed that along with Colletotrichum truncatum, several other species of Colletotrichum are associated with soybean worldwide, revealing a knowledge gap in what we know about this disease so far.

1. INTRODUCTION
The genus Colletotrichum constitutes a large monophyletic group of ascomycetes with more than 200 accepted species, classified into at least 14 species complexes (s.c.) and singletons (Damm et al., 2019; Marin‐Felix et al., 2017). Considered as among the top 10 plant pathogenic fungi, Colletotrichum spp. are the causal agents of anthracnose in more than 3,000 plant species, leading to considerable yield reduction of economically important crops (Cannon et al., 2012; Dean et al., 2012; da Silva et al., 2020). Due to its hemibiotrophic lifestyle and the facility of being manipulated in the laboratory, the genus is considered a model pathogen for biochemical, physiological, and genetic studies (Baroncelli et al., 2017; O’Connell et al., 2012; Perfect et al., 1999).
The soybean crop has great importance worldwide as a source of vegetable oil and proteins for human and animal feeds (Hartman et al., 2011; Pagano & Miransari, 2016), contributing 3.3% of the global human calorie intake (FAOSTAT, 2018). In 2019/20, world soybean production exceeded 330 million tonnes, of which approximately 86% were concentrated in Brazil, the United States, and Argentina (USDA, 2020). Diseases are among the major factors that can affect soybean production, and anthracnose is becoming a major threat in production areas around the world (Dias et al., 2016; Hartman et al., 2015; Nataraj et al., 2020; Subedi et al., 2016; Wrather et al., 2010;). This disease can reach up to 100% incidence in the field (Hartman et al., 1999), and incidence as low as 1% can cause yield losses of up to 90 kg/ha (Dias et al., 2016).
Soybean anthracnose is currently recognized as a disease of complex aetiology (Dias et al., 2018), with C. truncatum the most common associated species (Sharma et al., 2011). In recent years, several other species have been reported as causal agents of the disease, such as C. destructivum (Manandhar et al., 1986), C. coccodes (Riccioni et al., 1998), C. chlorophyti (Yang et al., 2012, 2013), C. gloeosporioides (Mahmodi et al., 2013), C. incanum (Yang et al., 2014), C. plurivorum (Barbieri et al., 2017), C. sojae (Damm et al., 2019), and, more recently, C. musicola (Boufleur et al., 2020) and C. brevisporum (Shi et al., 2020).
Colletotrichum species can affect soybean in all physiological stages (Sharma et al., 2011). Typical symptoms of anthracnose are pre‐ and postemergence damping‐off; and dark, depressed, and irregular spots on stems, petioles, and pods that can evolve and cause premature defoliation of the plants (Yang et al., 2015). Such infections have the potential to cause severe losses that can reach up to 100% in soybean‐producing areas under favourable conditions (Yang & Hartman, 2016).
An accurate species identification of a causal agent is decisive for disease control and prevention. The occurrence of multiple Colletotrichum species associated with soybean anthracnose may affect disease management because distinct species might respond differently to different control strategies. Different studies about the efficiency of fungicides in the control of soybean anthracnose showed contradictory results (Chen et al., 2018; Dias et al., 2016; Poti et al., 2020), which could be due to different responses of Colletotrichum species to their active compounds.
Few discriminatory morphological characters are available, and the identification of Colletotrichum species based exclusively on these features is unreliable (Cai et al., 2009; Jayawardena et al., 2016). Currently, species identification of this genus is performed using a polyphasic approach that combines morphological and cultural characteristics with multilocus phylogenetic analyses of DNA sequences (Cai et al., 2009; Liu et al., 2016).
Several species within the Colletotrichum genus show a wide genetic variability; however, the mechanisms responsible for such diversity are not yet fully understood (da Silva et al., 2020). Some population genetics studies of Colletotrichum species have offered tools for improving prevention and management strategies for plant diseases of important agricultural crops (Baroncelli et al., 2015; Ciampi‐Guillardi et al., 2014; Rogério et al., 2019; Ureña‐Padilla et al., 2002).
Many unresolved questions about soybean anthracnose remain. Until now, it has not been clear whether the association of new Colletotrichum species with the disease is related to emerging species or whether it is due to the ongoing changes in the taxonomy of the genus. Most of the studies available for soybean anthracnose are focused on C. truncatum, with limited information about the other Colletotrichum species infecting this crop, which could result in obstacles for the management of the disease in the field. The aim of this work is to gain a better understanding of soybean anthracnose and its causal agents, clarify which Colletotrichum species or complexes are associated with the disease, and provide a comprehensive review for future studies on soybean anthracnose.
2. COLLETOTRICHUM SPECIES ASSOCIATED WITH SOYBEAN, LIFESTYLE, AND GEOGRAPHIC DISTRIBUTION
The lack of reliable morphological characteristics has made the identification of Colletotrichum spp. a major challenge over the years (Cai et al., 2009), leading to considerable taxonomic confusion. After the advent of DNA‐based characterization, the taxonomy and nomenclature of Colletotrichum spp. underwent many revisions (Cannon et al., 2012; Damm et al., 2009, 2012a,2012b, 2014, 2019; Jayawardena et al., 2016; Liu et al., 2014; Marin‐Felix et al., 2017; Weir et al., 2012). Currently, there are more than 200 recognized Colletotrichum species, either as singletons or as part of 14 s.c. (Damm et al., 2019; Marin‐Felix et al., 2017). Classification of Colletotrichum into s.c. can be done using the internal transcribed spacer (ITS)‐5.8S rRNA region. A correct species identification requires a multilocus approach, with distinct s.c. demanding different loci to be analysed (Damm et al., 2019; Marin‐Felix et al., 2017).
Soybean anthracnose was first described in Korea in 1917, associated with C. glycines (Nakata & Takimoto, 1934). The same species was also reported as pathogenic to soybean in the USA in 1926 (Lehman & Wolf, 1926). Later, C. glycines (a synonym of C. truncatum) and Glomerella glycines were reported on soybean (Damm et al., 2019; Sharma et al., 2011; Sinclair, 1989). Until recently, studies of this disease have been mainly focused on C. truncatum. In contrast, the C. orchidearum s.c. was described only recently (Damm et al., 2019), and three species within this complex have already been reported as pathogenic to soybean (Barbieri et al., 2017; Boufleur et al., 2020; Lehman & Wolf, 1926). The recent massive taxonomic revision of this genus has led to an increase of taxonomically wrongly assigned ITS sequences deposited in GenBank (Crouch, Clarke, et al., 2009; Damm et al., 2009). To clarify the Colletotrichum s.c. associated with soybean anthracnose, and the worldwide distribution of this disease, all publicly available ITS sequences of Colletotrichum isolated from soybean and relative information were retrieved and compared with those of reference isolates through a phylogenetic analysis.
The in silico screening for Colletotrichum species isolated from soybean plants with and without symptoms whose nucleotide sequences are deposited in GenBank yielded 499 ITS sequences originally assigned to eight Colletotrichum s.c. (C. acutatum, C. boninense, C. dematium, C. gloeosporioides, C. magnum, C. orchidearum, C. spaethianum, and C. truncatum) and one singleton species (C. chlorophyti) (Table S1). While it has been shown that around 10% of all deposited ITS sequences are assigned to the wrong species (Nilsson et al., 2006), this number seems to be much higher in the case of Colletotrichum. In our data set more than 37% of the sequences were wrongly assigned at the s.c. level and therefore the proportion could be higher at the species level (Table S1). Our analyses suggest that Colletotrichum strains isolated from soybean belong to nine s.c. (C. acutatum, C. boninense, C. dematium, C. gloeosporioides, C. gigasporum, C. magnum, C. orchidearum, C. spaethianum, and C. truncatum) and one singleton species (C. chlorophyti) (Figure 1). To our knowledge this is the first time that species belonging to the C. gigasporum s.c. have been associated with soybean (Table 1).
FIGURE 1.
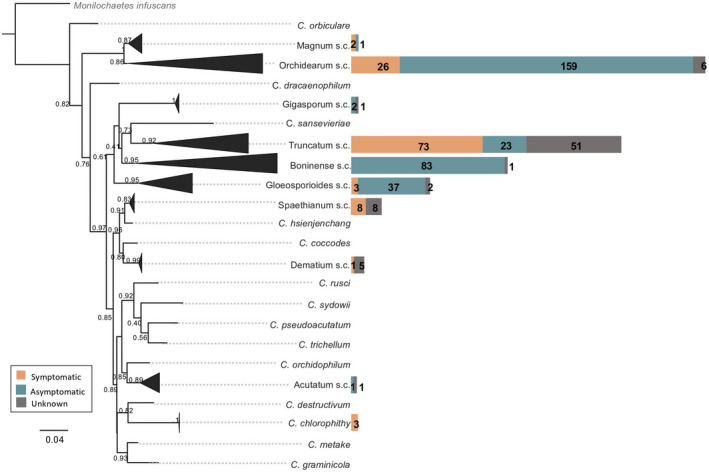
Colletotrichum species complexes (s.c.) associated with soybean worldwide identified based on a Bayesian phylogenetic analysis of internal transcribed spacer (ITS) sequences. Colletotrichum s.c. associated with symptomatic, asymptomatic, and unknown soybean plants are indicated by bars of distinct colours. All sequences were aligned using MAFFT v. 7.450 (Katoh, 2002; Katoh & Standley, 2013) and the multiple sequence alignment was exported to MEGA 10 (Stecher et al., 2020), in which the best‐fit substitution model was calculated for the sequence data set. The concatenated alignment was performed with Geneious v. 2020.0.4 (https://www.geneious.com). A Markov chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities with MrBayes v. 3.2.6 (Huelsenbeck & Ronquist, 2001) based on the model of nucleotide substitution. The analyses were run from random trees for 5,000,000 generations and sampled every 1,000 generations. The concatenated tree was compressed in FigTree v. 1.4.4 (Rambaut, 2014)
TABLE 1.
Colletotrichum species complexes associated with soybean anthracnose
| Species complex | Species | Distribution | Lifestyle | References |
|---|---|---|---|---|
| C. acutatum | Unknown | Brazil, USA | Endophyte | Leite et al., 2013 |
| C. boninense | Unknown 1 | Brazil, Taiwan | Endophyte | Leite et al., 2013 |
| C. gloeosporioides | C. gloeosporioides, C. salsolae | Brazil, Colombia, Hungary, Taiwan | Pathogen, endophyte | Weir et al., 2012; Leite et al., 2013; Chen et al., 2006 |
| C. spaethianum | C. incanum | Canada, USA | Pathogen | Yang et al., 2014 |
| C. gigasporum | Unknown 1 | Brazil | Endophyte | Leite et al., 2013 |
| C. dematium | Unknown 1 | Canada | Pathogen | Unpublished |
| None | C. chlorophyti | USA | Pathogen | Cannon et al., 2012; Yang et al., 2012; Yang et al., 2014 |
| C. magnum | C. brevisporum | Brazil, China, Taiwan | Pathogen, endophyte | Leite et al., 2013; Shi et al., 2020 |
| C. orchidearum | C. musicola, C. plurivorum, C. sojae | Brazil, Iran, Italy, Japan, Malasya, Myanmar, Serbia, Taiwan, USA | Pathogen, endophyte | Riccioni et al., 1998; Leite et al., 2013; Barbieri et al., 2017; Batzer & Muller, 2020 ; Damm et al., 2019; Boufleur et al., 2020; Zaw & Aye, 2020 |
| C. truncatum | C. truncatum | Brazil, Canada, China, Colombia, South Korea, Taiwan, USA | Pathogen, endophyte | Chen et al., 2006; Damm et al., 2009; Leite et al., 2013; Yang et al., 2014; Rogério et al., 2017; Zaw & Aye, 2020 |
No species could be assigned to the correct taxonomic position within the species complex due the lack of available information.
Although several s.c. have been associated with soybean (Figure 1), it was not possible to confirm if the strains belonging to the C. acutatum, C. boninense, and C. gigasporum s.c. are truly pathogenic to soybean, as all the sequences retrieved that belong to those complexes came from symptomless plants or information related to pathogenicity was not available. It is known that Colletotrichum can go through a quiescent phase before the development of disease symptoms on the host (Prusky, 1996; Prusky et al., 2013; de Silva et al., 2017) and can live inside nonhost plant tissues as endophytes (da Silva et al., 2020). Strains belonging to the C. dematium, C. magnum, C. gloeosporioides, C. orchidearum, C. truncatum s.c., and C. chlorophyti have been confirmed to be pathogenic (Table 1).
C. coccodes and C. destructivum were also reported to be pathogenic to soybean (Manandhar et al., 1986; Riccioni et al., 1998), but no genetic information is available and therefore we could not confirm the taxonomic designation of the strains used in those studies. C. coccodes is a singleton species isolated from soybean in the USA (Riccioni et al., 1998) and C. destructivum was reported for the first time as pathogenic to soybean in the 1980s, associated with the sexual morph G. glycines (Manandhar et al., 1986); thus, this species was recently reclassified as C. sojae (Damm et al., 2019). One of the most important soybean diseases in Argentina, the third major soybean producer of the world (USDA, 2020), is the late season disease complex that includes soybean anthracnose caused by C. truncatum and C. destructivum (Ramos et al., 2010, 2013). However, the real association of C. destructivum with the disease remains unclear, because those species were identified based only on morphological characteristics (Ramos et al., 2013).
Sixteen countries or regions around the world reported the presence of at least one Colletotrichum lineage associated with soybean. Brazil, the USA, and Taiwan have the largest diversity, followed by China, Colombia, Canada, and Myanmar (Figure 2). C. orchidearum and C. truncatum s.c. were the most widely distributed worldwide, present in nine countries, with the largest number of ITS sequences retrieved followed by the C. gloeosporioides s.c. present in four different countries (Figures 1 and 2), whereas the other s.c. seem to be restricted to one or two countries (Figure 2). Considering the number of records and the distribution of the C. orchidearum s.c. strains isolated since 2003 (Table S1) and their presence in several soybean‐producing countries, this complex is probably more important in the epidemiology of the disease than is currently assumed; thus, more studies need to be performed to understand its distribution, host specificity, and impact in soybean fields around the world.
FIGURE 2.
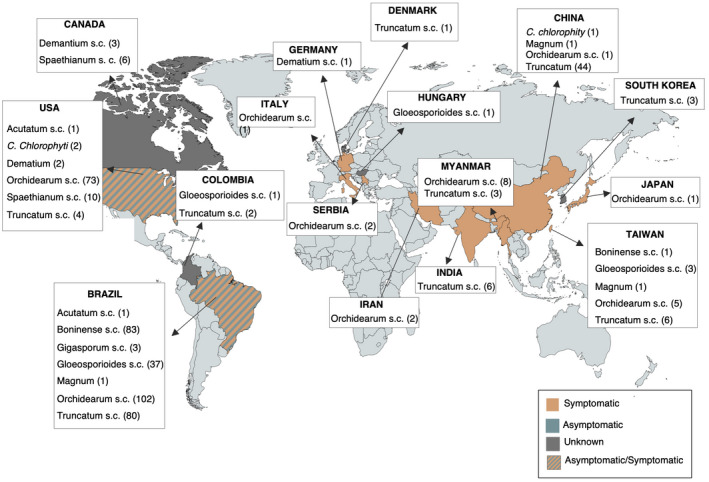
Global distribution of the Colletotrichum species complexes (s.c.) associated with soybean based on the information downloaded with sequences from GenBank. The number of isolates in each country or region is presented to the side of the names of the respective species complex. Countries or regions with the presence of symptomatic, asymptomatic, and unknown soybean plants are indicated by different colours
How many and which Colletotrichum species can be pathogenic or endophytic to soybean is still unclear. Historically, C. truncatum has been considered the prevalent species isolated from soybean or associated with soybean anthracnose (Sharma et al., 2011). However, many questions remain open, and based on the genetic data available (Table S1) we can hypothesize that the importance of C. truncatum has been overestimated. Precise knowledge about pathogen taxonomic designation and diversity is crucial, having direct implications on disease management, either by cultural or chemical strategies, as well as on disease resistance breeding programmes (Chen et al., 2018).
3. SYMPTOMATOLOGY AND EPIDEMIOLOGY
Favoured by warm and humid conditions, typical anthracnose symptoms can appear on all parts of soybean plants and in all physiological stages (Yang & Hartman, 2016). Systemic infections on seeds can cause pre‐ and/or postemergence damping‐off and cotyledon lesions (Sharma et al., 2011). Symptoms are commonly characterized by dark, depressed, and irregular spots on stems, petioles, and pods. Leaves may be shrunken, rolled or wilted, and may have necrotic laminar veins, resulting in premature defoliation of the plants (Figure 3) (Yang et al., 2015). This pattern of symptoms is reported for C. truncatum, C. coccodes, C. gloeosporioides, C. plurivorum, C. musicola, and C. incanum (Boufleur et al., 2020; Dias et al., 2018; Mahmodi et al., 2013; Riccioni et al., 1998; Sharma et al., 2011; Yang et al., 2014), while C. sojae causes circular to irregular greyish lesions with dark margins (Damm et al., 2019) and C. chlorophyti causes intra‐ and interveinal necrotic lesions surrounded by slight chlorosis (Yang et al., 2012).
FIGURE 3.

Typical symptoms and signs of Colletotrichum truncatum on infected soybean seeds (a–c). Dark, depressed, and irregular spots on cotyledons, stem, petioles, and pods (d–f)
Except for C. truncatum, almost no information is available on the life cycle of other Colletotrichum spp. associated with soybean anthracnose. C. truncatum survives on seeds, crop residues, and weeds, and can form soybean‐infective microsclerotia (Hartman et al., 1986; Khan & Sinclair, 1991; Yang & Hartman, 2016). Although the role of weeds and alternative hosts in the epidemiology of the disease is still unclear, probably the main source of primary inoculum of C. truncatum and C. plurivorum is infected seeds, which contribute to dispersion over long distances and the introduction of new fungal isolates in an area (Dias et al., 2018; Hartman et al., 1999).
Fungal penetration occurs directly after conidial germination and formation of an appressorium on the plant surface. C. truncatum and C. sojae have the same patterns of infection and colonization of soybean leaves (Manandhar et al., 1985). C. truncatum has a hemibiotrophic lifestyle with a first stage during which the penetration peg develops into a typical primary hypha that is a biotrophic vesicle between the cell wall and the plasma membrane. The biotrophic phase is followed by a switch to the necrotrophic phase, with the production of secondary hyphae that colonize the tissue intra‐ and intercellularly, causing cell death (Bhadauria et al., 2013). The possibility of quiescent/endophytic infection is also reported (Bhadauria et al., 2013); consequently plant tissues can be infected without showing any symptoms (Chen et al., 2006). Symptoms of anthracnose appear during the necrotrophic stage, in which the formation of acervuli containing conidia occurs. The conidia represent the secondary inoculum of the disease, disseminated by water splash that dissolves the mucilage in which they are covered, and aids short‐range dispersal (Madden, 1997). The life cycle of C. truncatum is illustrated in Figure 4.
FIGURE 4.
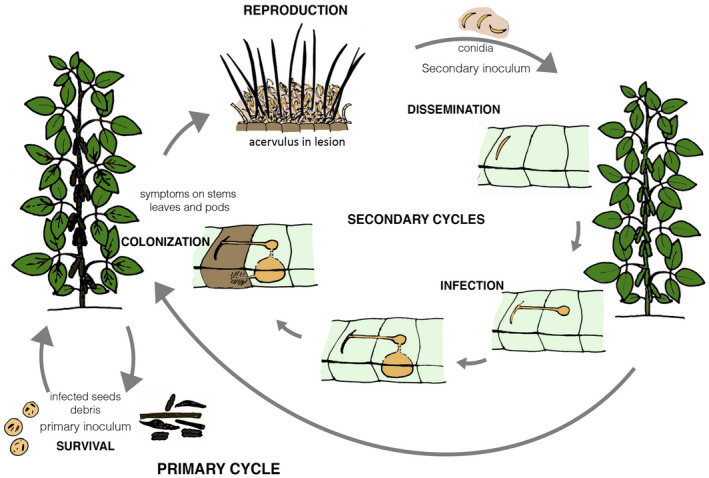
Disease cycle of soybean anthracnose caused by Colletotrichum truncatum
Understanding the relative importance of each s.c. in the development of the disease in the field is fundamental to direct epidemiological studies, which are essential for its effective management. Some species associated with soybean anthracnose, such as C. sojae, C. plurivorum, and C. musicola, have been reported to undergo the sexual state (Boufleur et al., 2020; Damm et al., 2019; Ramos et al., 2013). Glomerella glycines (now C. sojae) was considered the sexual morph of at least three species of Colletotrichum in the past, creating a taxonomic confusion that was solved by Damm et al. (2019), while others like C. truncatum and C. destructivum only occur in the asexual morph (Cannon et al., 2012; Damm et al., 2014). Can species with a sexual state have greater survivability on alternative hosts? Can genetic recombination play a role in survival or be an important source of variability in these species? Could sexual spores be spread differently from conidia? Do ascospores and conidia infect soybean tissues in the same way? These are some of the questions about the pathogen's epidemiology that still need answers as they directly impact the disease management.
4. DISEASE MANAGEMENT
The need for improvement of disease management motivated all efforts for a better understanding of soybean anthracnose proposed in this paper. The poor understanding of the life cycle and the epidemiological role of the Colletotrichum spp. associated with soybean may have led to ineffective disease management because it is not clear which species are responsible for the disease in the field.
Due to the potential for off‐season survival of species of Colletotrichum that infect soybean, and the long‐distance dissemination of the pathogen by seeds (Yang & Hartman, 2016), the management of soybean anthracnose should start with sowing disease‐free seeds (Pellegrino et al., 2010) and practising crop rotation. In most cases, seeds are symptomless; however, even low percentages of infection may lead to severe crop losses (Ciampi‐Guillardi et al., 2020; Pellegrino et al., 2010). To prevent the disease, seeds can be treated with systemic fungicides such as carboxanilide, dimethyldithiocarbamate, benzimidazoles, or triazoles (AGROFIT, 2020). Also, research on potential strategies of biological control with biopriming have been performed. Soybean seeds inoculated with Pseudomonas aeruginosa and Trichoderma harzianum reduced C. truncatum field incidence up to 92%, offering the same efficiency as the fungicide benomyl (Begum et al., 2010).
Currently, fungicides used as preventives are azoxystrobin, captan, mancozeb, carbendazim, thiophanate methyl, and members of the sterol demethylation inhibitors (DMI), such as triazoles (Dias et al., 2016; Nataraj et al., 2020; Poti et al., 2020). However, in recent years several studies have shown that fungicide efficiency is gradually reducing against soybean anthracnose (Dias et al., 2016; Poti et al., 2020).
In Brazil, when two seasons of soybean production were evaluated, chemical control with the use of triazoles combined with strobilurins was efficient during the first season, but not during the second season under natural Colletotrichum spp. infection (Dias et al., 2016). The resistance of C. truncatum isolates to multiple triazoles (flutriafol, fenbuconazole, tebuconazole, and metconazole) and reduced sensitivity to difenoconazole and propiconazole have been reported, indicating an inherent resistance as a result of CYP51A and CYP51B gene variations (Chen et al., 2018).
Carbendazim is a fungicide of the class of benzimidazole or methyl benzimidazole carbamate (MBC), which acts as a single‐site inhibitor (Oliver & Hewitt, 2014). Different studies showed that carbendazim is the most effective fungicide reducing C. truncatum growth in vitro (Agam et al., 2019; Ahamad et al., 2018; Kale & Barhate, 2016). During two seasons of soybean production in Nepal, in vivo trials showed that carbendazim (12%) combined with mancozeb (63%) reduced the disease incidence and increased the yield of the treated plots when compared with the control; this was considered the best treatment among the tested fungicides (Subedi et al., 2016). In contrast, in a study with 52 C. truncatum isolates from different fields, 89% of them were considered highly resistant to carbendazim (EC50 > 1,000 μg/ml) and 86% showed a mutation at codon 198 of the TUB2 gene, which prevents a hydrogen bond between carbendazim and β‐tubulin and is highly correlated with resistant fungal strains (Cai et al., 2015; Poti et al., 2020).
Although studies on the efficiency of fungicides against anthracnose in the field look promising, it remains unclear which Colletotrichum species are responsible for the disease. This may explain the contradictory results found in different field studies. The loss of efficacy has made producers intensify fungicide applications and doses, increasing their costs and inducing stronger selective pressure on the pathogen (Poti et al., 2020). The correct identification of the causal agent of anthracnose is important to explain the real reason for the lack of efficiency of the active compounds in the field.
Besides the potential of losses due to infection by Colletotrichum species (Wrather et al., 2010), there have been no breeding programmes for soybean cultivars resistant to anthracnose until now (Yang & Hartman, 2015). The implementation of resistant cultivars can generally reduce production costs and so this is a more eco‐friendly solution when compared with chemical control (Talhinhas et al., 2016).
Anthracnose resistance genes tend to be highly specific, and the emergence of additional Colletotrichum species associated with the disease in soybean‐producing areas indicates the need to start focused programmes (Dias et al., 2018). The inheritance of resistance to soybean anthracnose caused by C. truncatum was demonstrated to be governed by more than one gene (Nataraj et al., 2020). In a test with 16 soybean accessions inoculated with a mixture of C. truncatum isolates, Dias et al. (2019) found that some soybean genetic materials with a high level of resistance in stems are highly susceptible to cotyledon infection. They hypothesized that the genetic resistance of cotyledons and stems might be under the control of genetically independent mechanisms. Sources of resistance of soybean to anthracnose caused by C. truncatum have been reported and studies in Brazil, India, and the USA revealed that 22 commercial cultivars, nine genotypes, and one soybean germplasm are highly resistant to C. truncatum (Costa et al., 2009; Dias et al., 2019; Nagaraj et al., 2014, 2020; Yang & Hartman, 2015). Research on sources of resistance to other Colletotrichum species associated with anthracnose has not yet been performed.
5. IDENTIFICATION AND MOLECULAR DIAGNOSTICS
Accurate identification of Colletotrichum strains to the species level is critical in plant pathology with regard to fungal detection in propagative host material, quarantine measures, selection of biocontrol agents, screening varieties in plant breeding, population genetics, and genomics (Jayawardena et al., 2016). If cryptic species are confused with a single species, the integrity and understanding of the species will be compromised (Batista et al., 2017), and the importance of this is fundamental in population genetics and genomic studies.
5.1. Morphological characters
Most Colletotrichum lineages pathogenic to soybean can be divided into two major groups based on morphology: those that have curved conidia, including C. spaethianum, C. truncatum, and C. dematium s.c., plus C. chlorophyti (Damm et al., 2009); and those with straight cylindrical conidia, including C. gloeosporioides, C. gigasporum, C. magnum, and C. orchidearum s.c. (Damm et al., 2012a, 2012b, 2019; Liu et al., 2014; Weir et al., 2012). The main character of the C. acutatum s.c. is cylindrical conidia with acute ends (Damm et al., 2012a), while the C. boninense s.c. have straight cylindrical to clavate conidia (Damm et al., 2012b). Except for the C. gigasporum s.c., which have a distinctive morphological feature, with conidia up to 32 μm long and average length 26 μm (Liu et al., 2014), morphological characters overlap between species and s.c. associated with soybean, and have been fully described before (Damm et al., 2009, 2012a, 2012b, 2019; Liu et al., 2014; Weir et al., 2012); therefore, they should not be used for identification of Colletotrichum.
5.2. Molecular identification
Considering the importance of the Colletotrichum genus as a plant pathogen worldwide, rapid identification of a large collection of Colletotrichum isolates is often required. However, there is no consensus on the best molecular markers to discriminate species in each Colletotrichum s.c. (Vieira et al., 2020). Currently, there is no minimum or optimal standard set of molecular markers able to discriminate all the Colletotrichum s.c. (Marin‐Felix et al., 2017; Vieira et al., 2020). In general, five markers are amongst those commonly used to differentiate species among the distinct Colletotrichum s.c., especially those associated with soybean anthracnose: ITS, GAPDH, TUB2, CHS‐1, and ACT (Damm et al., 2009, 2019). Species within the C. acutatum s.c. can be effectively differentiated by both TUB2 and GAPDH markers (Damm et al., 2012a), while GAPDH alone can recognize all species within the C. boninense s.c. (Damm et al., 2012b). Combined gene analysis of ITS, GAPDH, CHS‐1, ACT, and TUB2 sequences can identify all the species within both the C. dematium and C. gigasporum s.c. (Liu et al., 2014). For the C. truncatum s.c. GAPDH is the most informative marker, followed by TUB2 and ACT (Vieira et al., 2020); for C. spaethianum and C. truncatum s.c., the inclusion of HIS3 in the multilocus combination is needed for the precise discrimination of species, whereas this locus is not informative for other complexes (Jayawardena et al., 2016). The combination of ITS, GAPDH, CHS‐1, HIS3, ACT, and TUB2 can differentiate species within the C. orchidearum s.c. (Damm et al., 2019). Species within the C. gloeosporioides s.c. can be distinguished by a combination of ApMat and GS sequences (Liu et al., 2015).
Because it might be unrealistic for most researchers to sequence multiple loci across a large set of isolates, it is useful to recommend markers with more phylogenetic informativeness (Vieira et al., 2020). In the majority of the 14 Colletotrichum species complexes GAPDH, HIS3, and TUB2 were found to be the most variable and informative markers for discriminating species.
The consensus is that species identification should not be based on BLAST searches of individual fungal sequences on NCBI/GenBank, but instead on robust phylogenetic analyses based on the concordance of multiple gene genealogies, including sequences from type species (Cannon et al., 2012). One reason is that most molecular markers alone do not exhibit sufficient polymorphism to discriminate Colletotrichum species, mainly within complexes, so that variation level among sequences is low. This is especially problematic in species with similar morphological characters that can be easily confused. Another issue is the problem of misidentification in the sequences deposited in NCBI as mentioned before, most likely as a consequence of the recent taxonomic reassessment of the genus that led to a massive increase in incorrectly assigned ITS sequences (Rogério et al., 2017). The ITS region should not be used singly to describe new Colletotrichum taxa because there is not enough discrimination for resolving the taxonomy.
Beyond phylogenetic trees, haplotype networks of concatenated sequences could be employed to infer geographical patterns of distribution or even host association among fungal lineages, below the species level. This approach was efficiently carried out in the identification of distinct clusters in C. truncatum strains causing soybean anthracnose in Brazil, by identifying groups of lineages associated with other Fabaceae hosts and weeds as well (Rogério et al., 2017).
5.3. Molecular diagnosis
In general, molecular diagnostic tests are developed on the basis of molecular markers largely used in phylogenetic studies. Despite some caveats, the most widely used molecular marker to design specific primers to detect fungal pathogens is the nuclear ribosomal cluster (Mancini et al., 2016; Pecchia et al., 2019). Recent studies have pointed out the lack of variation in the ITS region needed to develop specific primers for most Colletotrichum species (Da Lio et al., 2018). However, the intergenic spacer (IGS) region can be an alternative to ITS because it tends to contain more polymorphic sites and has proved to be an efficient marker for detecting C. lupini in lupins by PCR and could, therefore, be considered as an alternative target for other Colletotrichum species (Pecchia et al., 2019).
Molecular diagnostic techniques based on the detection of fungal DNA have been widely used for species‐specific detection of Colletotrichum associated with soybean anthracnose. PCR is the method of choice in the field of molecular diagnosis of soybean pathogens, as it enables an exponential amplification of the target DNA sequence, making it a fast, efficient, and attractive technique (Kumar et al., 2020). Using these molecular techniques, tiny amounts of host samples are sufficient for the detection of Colletotrichum in soybean seeds or other plant tissues. Several PCR‐based strategies are available for these purposes, such as multiplex PCR, loop‐mediated isothermal amplification (LAMP), real‐time or quantitative PCR (qPCR), and droplet digital PCR (ddPCR), among others, using specific primer pairs and sometimes excluding the need for DNA extraction (Ciampi‐Guillardi et al., 2020; Tian et al., 2017; Wang et al., 2017).
Multiplex qPCR assays have been consistently used for pathogen diagnosis in plant material by allowing the simultaneous amplification of multiple DNA targets in a single reaction (Schena et al., 2017). A highly sensitive multiplex TaqMan qPCR assay targeting the GAPDH gene was developed to detect and quantify as little as 0.3 pg of C. truncatum DNA, along with two other pathogens in soybean seeds (Ciampi‐Guillardi et al., 2020). The method was able to access fungal DNA directly from seed soaking solution, amplifying only the target species and not any other fungi commonly associated with soybean seeds. The high specificity of the assay is provided by the internal TaqMan probes, which overcomes the risk of false positives and/or false negatives. For the diagnosis of C. truncatum, a multiplex qPCR assay targeting the cox1 gene has also been proposed to distinguish four Colletotrichum species infecting soybean, C. chlorophyti, C. sojae, C. incanum, and C. truncatum, by using two duplex sets based on melting point temperatures. While successful detection was achieved with 0.1 pg of C. truncatum DNA, the assay may not be suitable for field diagnostics because it was tested only on purified Colletotrichum DNA, not on host tissue samples (Yang et al., 2015). The correct identification of Colletotrichum spp. in seeds is essential for diagnostic laboratories and producers, avoiding the introduction and dissemination of the pathogen in soybean fields (Ramiro et al., 2019). To date, there are still no diagnostic tests for all Colletotrichum species associated with soybean.
New tools have been developed to quickly detect Colletotrichum DNA in host samples. LAMP is a new nucleic acid amplification technology that enables the synthesis of large amounts of DNA in a short period of time with high specificity (Fu et al., 2011; Notomi et al., 2000). It could be a potential alternative to PCR because the LAMP protocol does not require a thermocycler. Despite the great potential attributed to the technique, LAMP has not been widely used for detecting Colletotrichum species associated with soybean anthracnose so far. Rapid LAMP diagnostic assays were proposed to detect C. truncatum, targeting the large subunit of RNA polymerase II (Rpb1) coding gene (Tian et al., 2017), and C. gloeosporioides, whose target was a glutamine synthetase (GS) gene (Wang et al., 2017) in soybean samples. For C. truncatum the detection limit of the LAMP assay was 100 pg/μl of fungal DNA per reaction, a hundred times greater than the amount detected in the qPCR assay proposed by Tian et al. (2017) and more than a thousand times less sensitive than the qPCR assay developed by Ciampi‐Guillardi et al. (2020).
An alternative approach would be to identify genomic regions specific to emerging Colletotrichum species or even to particular lineages using a computational approach based on whole‐genome comparison of distinct isolates or lineages. This approach has been successfully used to develop specific markers for the detection of C. lupini and other plant pathogens (Pecchia et al., 2019; Pieck et al., 2017; Thierry et al., 2020). Specific care must be taken in cases of recently diverged taxa, bearing in mind that it is unlikely that a single genomic region would perfectly meet all the requirements of a specific detection, especially in fungal lineages with very low divergence levels and recent genetic exchanges between them (Thierry et al., 2020).
6. POPULATION GENETIC STUDIES
Genetic variability in the form of the presence of different alleles occurring at different frequencies in genes is crucial to provide greater endurance to environmental changes and to increase species local adaptation over time (Barrett & Schluter, 2008; Hartl & Clark, 1997). Genetic investigation using a group of individuals provides a broader overview of species variability than a study with few individuals (McDonald, 1997) and can be used to make inferences about the predominant mode of pathogen reproduction and their impact on genotypic diversity (McDonald & Linde, 2002). In recent years, plant pathologists have been interested in investigations of genetic variation in pathogen populations, providing tremendous insights into the biology of fungal plant parasites (Giraud et al., 2008).
Knowledge of genetic structure, that is, the amount and distribution of genetic variation within and among populations, allows us to investigate the evolutionary forces (gene flow, genetic drift, mutation, and natural selection) acting as modulators of genetic diversity in populations (Giraud et al., 2008). The evolutionary potential of pathogen populations is directly guided by their genetic diversity (Croll & Laine, 2016). Thus, knowledge of the genetic structure gives information about the evolutionary processes that influenced plant pathogen populations in the past and provides insights into their future evolutionary potential (McDonald & Linde, 2002). Such information could be useful to optimize the management of resistance genes and fungicides in agriculture, and therefore to control plant diseases more effectively (Zhan, 2009).
Several population genetic studies of Colletotrichum species have been published over the years, and these investigations have increased our knowledge of the genetic variation of many important agricultural species (Banniza et al., 2018; Baroncelli et al., 2015; Ciampi‐Guillardi et al., 2014; Crouch, Tredway, et al., 2009; Ureña‐Padilla et al., 2002; Xavier et al., 2018). Despite the importance of soybean anthracnose, few studies are available on this pathosystem, and population genetic studies are even more scarce. Previous research mainly focused on genetic differences among C. truncatum isolates obtained from a range of hosts using distinct types of genetic markers, which revealed high genetic diversity and possible genetic recombination (Ford et al., 2004; Katoch et al., 2017; Ranathunge et al., 2009; Rogério et al., 2017; Sant’anna et al., 2010; Sharma, 2009; Vasconcelos et al., 1994;).
As C. truncatum was the only fungal species associated with soybean anthracnose in Brazil up to 2007 (Rogério et al., 2017), an investigation of the genetic structure of C. truncatum populations in the main soybean production areas was performed (Rogério et al., 2019). High levels of genetic diversity within populations and no evidence of intraregional gene flow were revealed by microsatellite data. This study also suggested that Brazilian C. truncatum populations resulted from at least three founder events, which led to three genetic groups that spread throughout the country, conserving syntopy (Rogério et al., 2019). Another study investigated populations of C. truncatum from Brazil and Argentina and identified that intragroup similarity was greater among the Argentinian isolates than the Brazilian group, with a strong correlation between geographical origin and genetic grouping (Dias et al., 2019). Such a large difference in genetic diversity was associated with a greater geographic breadth of the sampling in Brazilian populations, in addition to a greater genetic variability of host cultivars, which could be reflected in the variability of the isolates among Brazilian regions.
Considering the increase of soybean anthracnose in South America, population studies suggest an association between the inherent variability of the pathogen and the climatic and cultural features, as well the genetic makeup of commercial soybean cultivars used (Dias et al., 2019; Rogério et al., 2019). These population studies highlight intraspecific pathogen variability as a major feature in genetic breeding for anthracnose resistance. Efforts in breeding programmes aiming at anthracnose resistance should take into account the population structure and the genetic diversity levels of the pathogen by using representative isolates of the genetic variability of the species for screening soybean resistant cultivars.
Although there have been advances in understanding the genetic variation in C. truncatum infecting soybean and its impact on disease management strategies, many gaps have not yet been filled. Expanding the discrimination of genetic groups recently detected and the estimation of recombination rates may provide a potent approach to elucidate the pathogen life history and to address fundamental questions about the evolution and demographic history of this species (Rogério et al., 2019; Stukenbrock, 2016).
7. GENOMICS AS A TOOL FOR UNDERSTANDING PATHOGENICITY FACTORS
Recent technological advances in next‐generation sequencing and computational tools have made it possible to sequence and analyse whole genomes of many plant pathogens (Sant’Anna et al., 2010). These technologies continue to advance rapidly, and costs have declined to the point that it is becoming affordable to sequence genomes of many individuals within a species (Grünwald et al., 2016; Raffaele & Kamoun, 2012). This genomic revolution provides a major opportunity to connect the gaps between molecular biology, evolutionary genetics, and epidemiology (Plissonneau et al., 2017), playing a key role in plant disease management strategies (Klosterman et al., 2016).
The availability of a large number of genetic markers distributed throughout the genome enables the refinement of molecular variation investigations. The use of these markers provides fine‐grained resolution of genetic divergence, recombination, demography, as well as evolutionary biology of pathogen populations, enabling more robust inferences compared to studies based on a limited number of genetic markers (Brumfield et al., 2003; Helyar et al., 2011; Luikart et al., 2003). Population genomics analyses of a large number of loci offer an excellent opportunity to determine the genetic basis of many fungal phenotypes, including virulence (Plissonneau et al., 2017; Sarrocco et al., 2020). Furthermore, techniques such as genome‐wide association studies (GWAS), quantitative trait locus (QTL) mapping, and genome scans for signatures of selection and selective sweeps are powerful tools to identify genes involved in host‐specific interactions of fungal pathogens (Grünwald et al., 2016; Plissonneau et al., 2017).
To date, four Colletotrichum genomes isolated from soybean plants with symptoms have been sequenced: C. truncatum (IMI 507125), C. plurivorum (IMI 507127), C. musicola (IMI 507128), and C. sojae (IMI 507126) (Rogério et al., 2020). Additionally, the genome of two other strains of C. truncatum, MTCC 3114 and TYU, isolated from Capsicum annuum and Taxus cuspidata, respectively, and a strain of C. chlorophyti (NTL11) from tomato (Solanum lycopersicon) are currently available (Gan et al., 2017; Rao & Nandineni, 2017; Rogério et al., 2020). A summary of the genome assembly statistics is available in Table 2. The pathogenicity of the four Colletotrichum spp. sequenced by Rogério et al. (2020) to soybean fulfilled Koch's postulates on soybean. While the other sequenced isolates (MTCC 3114, TYU, and NTL11) belong to species pathogenic to soybean, it was not confirmed if they can infect soybean. The genome sequence data of Colletotrichum species pathogenic to soybean currently available may greatly aid our understanding of host–pathogen interactions besides offering a useful resource for further research in comparative genomics and evolutionary studies of Colletotrichum.
TABLE 2.
Summary of Colletotrichum spp. pathogenic to soybean that have whole genome sequences
| Species | Strain | Host | Country | Accession no. | Assembly length (Mb) | GC (%) | No. of predicted genes | Reference |
|---|---|---|---|---|---|---|---|---|
| C. chlorophyti | NTL11 | Solanum lycopersicon | Japan | MPGH00000000 | 52.40 | 50.06 | 10,419 | Gan et al., 2017 |
| C. musicola | IMI 507128 | Glycine max | Brazil | WIGM00000000 | 52.73 | 54.97 | 16,826 | Rogério et al., 2020 |
| C. plurivorum | IMI 507127 | G. max | Brazil | WIGO00000000 | 49.70 | 55.86 | 16,153 | Rogério et al., 2020 |
| C. sojae | IMI 507126 | G. max | Brazil | WIGN00000000 | 49.35 | 55.92 | 16,124 | Rogério et al., 2020 |
| C. truncatum | IMI 507125 | G. max | Brazil | VUJX00000000 | 56.10 | 50.12 | 15,901 | Rogério et al., 2020 |
| C. truncatum | MTCC 3114 | Capsicum annum | India | NBAU00000000 | 55.30 | 49.61 | 13,724 | Rao & Nandineni, 2017 |
| C. truncatum | TYU | Taxus cuspidata | South Korea | NOWE00000000 | 53.00 | 49.61 | – | – |
| C. truncatum | GO2−03 | G. max | Brazil | SRX7095355 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT5−32 | G. max | Brazil | SRX7095354 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT5−26 | G. max | Brazil | SRX7095353 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT5−12 | G. max | Brazil | SRX7095352 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT4−13 | G. max | Brazil | SRX7095351 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT4−05 | G. max | Brazil | SRX7095350 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT3−21 | G. max | Brazil | SRX7095349 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT3−01 | G. max | Brazil | SRX7095348 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO5−25 | G. max | Brazil | SRX7095347 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO5−14 | G. max | Brazil | SRX7095346 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO5−11 | G. max | Brazil | SRX7095345 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO4−17 | G. max | Brazil | SRX7095344 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO4−08 | G. max | Brazil | SRX7095343 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO4−07 | G. max | Brazil | SRX7095342 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO2−12 | G. max | Brazil | SRX7095341 | – | – | – | Rogério et al., 2019 |
| C. truncatum | GO2−06 | G. max | Brazil | SRX7095340 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT2−05 | G. max | Brazil | SRX7095339 | – | – | – | Rogério et al., 2019 |
| C. truncatum | MT1−01 | G. max | Brazil | SRX7095338 | – | – | – | Rogério et al., 2019 |
Scanning the Colletotrichum genomes for identification of full putative effector repertoires of the pathogen may be a useful tool for soybean breeders in the development of new cultivars with durable resistance against anthracnose (Barsoum et al., 2019; Lenman et al., 2016; Oliver & Solomon, 2010; Prasad et al., 2019; Van de Wouw & Idnurm, 2019). Although there is no breeding programme aimed at screening resistance to soybean anthracnose so far, the genomic resources now available may support the development of future programmes. Additionally, genome resources can be used to accelerate the development of diagnostic tools for plant pathogens (Klosterman et al., 2016), impacting the application of more accurate management strategies.
Advances in comparative genomics and population genomic approaches open new perspectives to increase our understanding of the molecular mechanisms underpinning the pathogenesis and adaptive processes of these pathogens (Klosterman et al., 2016; Stukenbrock & Bataillon, 2012). Mechanisms regarding C. truncatum pathogenicity have been investigated to understand the factors involved in the pathogenesis of different hosts (Auyong, 2015; Auyong et al., 2012; Madden, 1997; Ranathunge et al., 2009). With the availability of genomic sequences for Colletotrichum species pathogenic to soybean, new advances into pathogenic processes at the molecular level are possible, contributing to improving our knowledge in the host–fungal interactions in the soybean–anthracnose pathosystem, and thus developing effective and novel strategies to combat the pathogens.
8. CONCLUSIONS AND FUTURE PERSPECTIVES
Knowing precisely the diversity of a pathogen is crucial from taxonomic, biological, and ecological standpoints. Indeed, pathogen identity has direct implications for disease management by cultural or chemical strategies as well as for disease resistance breeding programmes. Furthermore, an effective management of new Colletotrichum species requires tools to discriminate between emerging and established fungal populations associated with soybean, aiming to detect the pathogens at the earliest point to monitor and limit their spread.
C. truncatum has been considered the most important causal agent of soybean anthracnose. However, our survey showed that at least 12 Colletotrichum lineages are associated with soybean, with the C. truncatum and C. orchidearum s.c. having the greatest impact and the broadest worldwide distribution. Most of the information available on soybean anthracnose until now has been limited to C. truncatum. Taking into account the numerous Colletotrichum species causing the disease, there is a gap in the knowledge of epidemiology, worldwide movement, distribution, identification, control measures, fungicide efficiency, and genetic resistance for all of the species.
In agreement with Vieira et al. (2020), more robust genomic sampling is required to improve our understanding of relationships among taxa in the genus Colletotrichum, and also our ability to distinguish species within s.c. Genome data is now available for several Colletotrichum species associated with anthracnose in soybean, such as C. truncatum, C. musicola, C. plurivorum, C. sojae, and C. chlorophyti (Gan et al., 2017; Rao & Nandineni, 2017; Rogério et al., 2020), yet a comprehensive phylogenomic study of the genus is still needed. A population genomics approach and comparative genomics investigations can be used to identify candidate genes involved in pathogenicity, virulence (or aggressiveness), host specialization, fungicide resistance, and adaptation to different environments with higher precision, contributing to a better understanding of Colletotrichum species dynamic in the agroecosystems.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Supporting information
TABLE S1 Strains of Colletotrichum spp. studied, with collection details and GenBank accession numbers
ACKNOWLEDGEMENTS
This research was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo, grant number 2017/09178‐8, the National Science and Technology Development Council, grant numbers 140668/2018‐7 and 142376/2020‐5, and the National Council for the Improvement of Higher Education, CAPES/PDSE grant number 88881.133223/2016‐01, PROEX/CAPES grant number 455 330002037002P3, and the Ministerio de Ciencia Innovación y Universidades of Spain, grant number RTI2018‐093611‐B‐I00.
Boufleur TR, Ciampi‐Guillardi M, Tikami Í, et al. Soybean anthracnose caused by Colletotrichum species: Current status and future prospects. Mol Plant Pathol. 2021;22:393–409. 10.1111/mpp.13036
DATA AVAILABILITY STATEMENT
Data sharing is not applicable as no new data were generated or analysed.
REFERENCES
- Agam, M.N. , Raut, R.A. , Jejurkar, G.B. & Sable, S.B. (2019) Evaluation of the fungicides, botanicals and bioagents against Colletotrichum truncatum causing anthracnose of soybean in pot culture. Journal of Pharmacognosy and Phytochemistry, 8, 629–634. [Google Scholar]
- AGROFIT (2020). Sistemas de agrotóxicos fitossanitários. Available at: http://extranet.agricultura.gov.br/agrofit_cons/principal_agrofit_cons [accessed August 2020].
- Ahamad, V. , Kaintura, P. & Gaur, G. (2018) In vitro evaluation of fungicides and bio‐control agents against Colletotrichum truncatum in soybean in Dehradun Valley. International Journal of Pure and Applied Bioscience, 6, 869–874. [Google Scholar]
- Auyong, A.S. (2015) The role of cutinase and its impact on pathogenicity of Colletotrichum truncatum . Journal of Plant Pathology and Microbiology, 6, 1000259. [Google Scholar]
- Auyong, A.S.M. , Ford, R. & Taylor, P.W.J. (2012) Genetic transformation of Colletotrichum truncatum associated with anthracnose disease of chili by random insertional mutagenesis. Journal of Basic Microbiology, 52, 372–382. [DOI] [PubMed] [Google Scholar]
- Banniza, S. , Warale, R. , Menat, J. , Cohen‐Skali, A. , Armstrong‐Cho, C. & Bhadauria, V. (2018) The long path to understanding the host–pathogen interactions of Colletotrichum lentis on lentil. Canadian Journal of Plant Pathology, 40, 199–209. [Google Scholar]
- Barbieri, M.C.G. , Ciampi‐Guillardi, M. , Moraes, S.R.G. , Bonaldo, S.M. , Rogério, F. , Linhares, R.R. et al. (2017) First report of Colletotrichum cliviae causing anthracnose on soybean in Brazil. Plant Disease, 101, 1677. [Google Scholar]
- Baroncelli, R. , Talhinhas, P. , Pensec, F. , Sukno, S.A. , Le Floch, G. & Thon, M.R. (2017) The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Frontiers in Microbiology, 8, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroncelli, R. , Zapparata, A. , Sarrocco, S. , Sukno, S.A. , Lane, C.R. , Thon, M.R. et al. (2015) Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species . PLoS One, 10, e0129140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, R. & Schluter, D. (2008) Adaptation from standing genetic variation. Trends in Ecology and Evolution, 23, 38–44. [DOI] [PubMed] [Google Scholar]
- Barsoum, M. , Sabelleck, B. , Spanu, D.P. & Panstruga, R. (2019) Rumble in the effector jungle: Candidate effector proteins in interactions of plants with powdery mildew and rust fungi. Critical Reviews in Plant Sciences, 38, 255–279. [Google Scholar]
- Batista, D. , Silva, D.N. , Vieira, A. , Cabral, A. , Pires, A.S. , Loureiro, A. et al. (2017) Legitimacy and implications of reducing Colletotrichum kahawae to subspecies in plant pathology. Frontiers in Plant Science, 7, 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, J.C. & Mueller, D.S. (2020) Soybean fungal endophytes Alternaria and Diaporthe spp. are differentially impacted by fungicide application. Plant Disease, 104, 52–59. [DOI] [PubMed] [Google Scholar]
- Begum, M.M. , Sariah, M. , Puteh, A.B. , Zainal Abidin, M.A. , Rahman, M.A. & Siddiqui, Y. (2010) Field performance of bio‐primed seeds to suppress Colletotrichum truncatum causing damping‐off and seedling stand of soybean. Biological Control, 53, 18–23. [Google Scholar]
- Bhadauria, V. , Banniza, S. , Vandenberg, A. , Selvaraj, G. & Wei, Y. (2013) Overexpression of a novel biotrophy‐specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryotic Cell, 12, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boufleur, T.R. , Castro, R.R.L. , Rogério, F. , Ciampi‐Guillardi, M. , Baroncelli, R. & Massola Júnior, N.S. (2020) First report of Colletotrichum musicola causing soybean anthracnose in Brazil. Plant Disease, 104, 1858. [DOI] [PubMed] [Google Scholar]
- Brumfield, R.T. , Beerli, P. , Nickerson, D.A. & Edwards, S.V. (2003) The utility of single nucleotide polymorphisms in inferences of population history. Trends in Ecology and Evolution, 18, 249–256. [Google Scholar]
- Cai, L. , Hyde, K.D. , Taylor, P.W.J. , Weir, B.S. , Waller, J.M. , Abang, M.M. et al. (2009) A polyphasic approach for studying Colletotrichum . Fungal Diversity, 39, 183–204. [Google Scholar]
- Cai, M. , Lin, D. , Chen, L. , Bi, Y. , Xiao, L. & Liu, X. (2015) M233I mutation in the β‐tubulin of Botrytis cinerea confers resistance to zoxamide. Scientific Reports, 5, 16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, P.F. , Damm, U. , Johnston, P.R. & Weir, B.S. (2012) Colletotrichum – current status and future directions. Studies in Mycology, 73, 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.S. , Chu, C. , Liu, C.D. , Chen, R.S. & Tsay, J.G. (2006) PCR‐based detection and differentiation of anthracnose pathogens, Colletotrichum gloeosporioides and C. truncatum, from vegetable soybean in Taiwan: PCR‐based detection of Glomerella/Colletotrichum spp. Journal of Phytopathology, 154, 654–662. [Google Scholar]
- Chen, S. , Wang, Y. , Schnabel, G. , Peng, C.A. , Lagishetty, S. , Smith, K. et al. (2018) Inherent resistance to 14α‐demethylation inhibitor fungicides in Colletotrichum truncatum is likely linked to CYP51A and/or CYP51B gene variants. Phytopathology, 108, 1263–1275. [DOI] [PubMed] [Google Scholar]
- Ciampi‐Guillardi, M. , Baldauf, C. , Souza, A.P. , Silva‐Junior, G.J. & Amorim, L. (2014) Recent introduction and recombination in Colletotrichum acutatum populations associated with citrus postbloom fruit drop epidemics in São Paulo, Brazil. Phytopathology, 104, 769–778. [DOI] [PubMed] [Google Scholar]
- Ciampi‐Guillardi, M. , Ramiro, J. , de Moraes, M.H.D. , Barbieri, M.C.G. & Massola, N.S. (2020) Multiplex qPCR assay for direct detection and quantification of Colletotrichum truncatum, Corynespora cassiicola, and Sclerotinia sclerotiorum in soybean seeds. Plant Disease, 104, 3002–3009. [DOI] [PubMed] [Google Scholar]
- Costa, I.F.D. , Balardin, R.S. , Medeiros, L.A.M. , Lenz, G. , Gulart, C.A. , Zemolin, C.R. et al. (2009) Reação de germoplasma comercial de soja a Colletotrichum truncatum . Tropical Plant Pathology, 34, 47–50. [Google Scholar]
- Croll, D. & Laine, A.‐L. (2016) What the population genetic structures of host and pathogen tell us about disease evolution. New Phytologist, 212, 537–539. [DOI] [PubMed] [Google Scholar]
- Crouch, J.A. , Clarke, B.B. & Hillman, B.I. (2009) What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate‐spored graminicolous Colletotrichum group. Mycologia, 101, 648–656. [DOI] [PubMed] [Google Scholar]
- Crouch, J.A. , Tredway, L.P. , Clarke, B.B. & Hillman, B.I. (2009) Phylogenetic and population genetic divergence correspond with habitat for the pathogen Colletotrichum cereale and allied taxa across diverse grass communities. Molecular Ecology, 18, 123–135. [DOI] [PubMed] [Google Scholar]
- Da Lio, D. , Cobo‐Díaz, J.F. , Masson, C. , Chalopin, M. , Kebe, D. , Giraud, M. et al. (2018) Combined metabarcoding and multi‐locus approach for genetic characterization of Colletotrichum species associated with common walnut (Juglans regia) anthracnose in France. Scientific Reports, 8, 10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm, U. , Cannon, P.F. , Woudenberg, J.H.C. & Crous, P.W. (2012a) The Colletotrichum acutatum species complex. Studies in Mycology, 73, 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm, U. , Cannon, P.F. , Woudenberg, J. , Johnston, P.R. , Weir, B.S. , Tan, Y.P. et al. (2012b) The Colletotrichum boninense species complex. Studies in Mycology, 73, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm, U. , O’Connell, R.J. , Groenewald, J.Z. & Crous, P.W. (2014) The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology, 79, 49–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm, U. , Sato, T. , Alizadeh, A. , Groenewald, J.Z. & Crous, P.W. (2019) The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Studies in Mycology, 92, 1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm, U. , Wounderberg, J.H.C. , Cannon, P.F. & Crous, P.W. (2009) Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity, 39, 45–87. [Google Scholar]
- Dean, R. , van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , di Pietro, A. , Spanu, P.D. et al. (2012) The Top 10 fungal pathogens in molecular plant pathology: Top 10 fungal pathogens. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, M.D. , Dias‐Neto, J.J. , Santos, M.D.M. , Formento, A.N. , Bizerra, L.V.A.S. , Fonseca, M.E.N. et al. (2019) Current status of soybean anthracnose associated with Colletotrichum truncatum in Brazil and Argentina. Plants, 8, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, M.D. , Fonseca, M.E.N. , Dias‐Neto, J.J. , Santos, M.D.M. , Pandolfo, G.M. , Boiteux, L.S. et al. (2018) Biology, pathogenicity, and haplotype analyses of Colletotrichum cliviae: A novel soybean anthracnose agent in warm tropical areas. Tropical Plant Pathology, 43, 439–451. [Google Scholar]
- Dias, M.D. , Pinheiro, V.F. & Café‐Filho, A.C. (2016) Impact of anthracnose on the yield of soybean subjected to chemical control in the north region of Brazil. Summa Phytopathologica, 42, 18–23. [Google Scholar]
- FAOSTAT (Food and Agriculture Organization of the United Nations) (2018) Food and agriculture data. Available at: http://www.fao.org/faostat/en [accessed August 2020].
- Ford, R. , Banniza, S. , Phaotita, W. & Taylor, P.W.J. (2004) Morphological and molecular discrimination of Colletotrichum truncatum causing anthracnose on lentil in Canada. Australasian Plant Pathology, 33, 559–569. [Google Scholar]
- Fu, S. , Qu, G. , Guo, S. , Ma, L. , Zhang, N.A. , Zhang, S. et al. (2011) Applications of loop‐mediated isothermal DNA amplification. Applied Biochemistry and Biotechnology, 163, 845–850. [DOI] [PubMed] [Google Scholar]
- Gan, P. , Narusaka, M. , Tsushima, A. , Narusaka, Y. , Takano, Y. & Shirasu, K. (2017) Draft genome assembly of Colletotrichum chlorophyti, a pathogen of herbaceous plants. Genome Announcements, 5, e01733–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud, T. , Enjalbert, J. , Fournier, E. , Delmotte, F. & Dutech, C. (2008) Population genetics of fungal diseases of plants. Parasite, 15, 449–454. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. , McDonald, B.A. & Milgroom, M.G. (2016) Population genomics of fungal and oomycete pathogens. Annual Review of Phytopathology, 54, 323–346. [DOI] [PubMed] [Google Scholar]
- Hartl, D.L. & Clark, A.G. (1997) Principles of Population Genetics, 3rd edition. Sinauer Associates Inc. [Google Scholar]
- Hartman, G.L. , Bowen, C.R. , Haudenshield, J.S. , Fox, C.M. , Cary, T.R. & Diers, B.W. (2015) Evaluation of disease and pest damage on soybean cultivars released from 1923 through 2008 under field conditions in central Illinois. Agronomy Journal, 107, 2373–2380. [Google Scholar]
- Hartman, G.L. , Manandhar, J.B. & Sinclair, J.B. (1986) Incidence of Colletotrichum spp. on soybean and weeds in Illinois and pathogenicity of Colletotrichum truncatum . Phytopathology, 70, 780–782. [Google Scholar]
- Hartman, G.L. , Sinclair, J.B. & Rupe, J.C. (1999) Compendium of soybean diseasses, 4th edition. APS Press. [Google Scholar]
- Hartman, G.L. , West, E.D. & Herman, T.K. (2011) Crops that feed the world 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Security, 3, 5–17. [Google Scholar]
- Helyar, S.J. , Hemmer‐Hansen, J. , Bekkevold, D. , Taylor, M.I. , Ogden, R. , Limborg, M.T. et al. (2011) Application of SNPs for population genetics of nonmodel organisms: New opportunities and challenges: Analytical approaches. Molecular Ecology Resources, 11, 123–136. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- Jayawardena, R.S. , Hyde, K.D. , Damm, U. , Cai, L. , Liu, M. , Li, X.H. , Zhang, W. , Zhao, W.S. & Yan, J.Y. (2016) Notes on currently accepted species of Colletotrichum . Mycosphere, 7, 1192–1260. [Google Scholar]
- Kale, S.L. & Barhate, B.G. (2016) Management of anthracnose in soybean caused by Colletotrichum truncatum . International Journal of Plant Protection, 9, 583–588. [Google Scholar]
- Katoch, A. , Sharma, P. , Padder, B.A. & Sharma, P.N. (2017) Population structure of Colletotrichum truncatum in Himachal Pradesh and identification of broad‐spectrum resistant sources in Capsicum . Agricultural Research, 6, 296–303. [Google Scholar]
- Katoh, K. (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. & Sinclair, J.B. (1991) Effect of soil temperature on infection of soybean roots by sclerotia‐forming isolates of Colletotrichum truncatum . Plant Disease, 75, 1282–1285. [Google Scholar]
- Klosterman, S.J. , Rollins, J.R. , Sudarshana, M.R. & Vinatzer, B.A. (2016) Disease management in the genomics era—summaries of focus issue papers. Phytopathology, 106, 1068–1070. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Gupta, A. , Srivastava, S. , Devi, G. , Singh, V.K. , Goswami, S.K. et al. (2020) Diagnosis and detection of seed‐borne fungal phytopathogens. In: Kumar, R. and Gupta, A. (Eds.) Seed‐Borne Diseases of Agricultural Crops: Detection, Diagnosis and Management. : Springer Singapore, pp. 107–142. [Google Scholar]
- Lehman, S.G. & Wolf, F.A. (1926) Soybean anthracnose. Journal of Agricultural Research, 33, 381–390. [Google Scholar]
- Leite, T.S. , Cnossen‐Fassoni, A. , Pereira, O.L. , Mizubuti, E.S.G. , Araújo, E.F. & Queiroz, M.V. (2013) Novel and highly diverse fungal endophytes in soybean revealed by the consortium of two different techniques. Journal of Microbiology, 51, 56–69. [DOI] [PubMed] [Google Scholar]
- Lenman, M. , Ali, A. , Mühlenbock, P. , Carlson‐Nilsson, U. , Liljeroth, E. , Champouret, N. et al. (2016) Effector‐driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93‐1015. Theoretical and Applied Genetics, 129, 105–115. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Cai, L. , Crous, P.W. & Damm, U. (2014) The Colletotrichum gigasporum species complex. Persoonia, 33, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Wang, M. , Damm, U. , Crous, P.W. & Cai, L. (2016) Species boundaries in plant pathogenic fungi: A Colletotrichum case study. BMC Evolutionary Biology, 16, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Weir, B.S. , Damm, U. , Crous, P.W. , Wang, Y. , Liu, B. et al. (2015) Unravelling Colletotrichum species associated with Camellia: Employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia, 35, 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart, G. , England, P.R. , Tallmon, D. , Jordan, S. & Taberlet, P. (2003) The power and promise of population genomics: From genotyping to genome typing. Nature Reviews Genetics, 4, 981–994. [DOI] [PubMed] [Google Scholar]
- Madden, L.V. (1997) Effects of rain on splash dispersal of fungal pathogens. Canadian Journal of Plant Pathology, 19, 225–230. [Google Scholar]
- Mahmodi, F. , Kadir, J.B. , Wong, M.Y. , Nasehi, A. , Puteh, A. & Soleimani, N. (2013) First report of anthracnose caused by Colletotrichum gloeosporioides on soybean (Glycine max) in Malaysia. Plant Disease, 97, 841. [DOI] [PubMed] [Google Scholar]
- Manandhar, J.B. , Hartman, G.L. & Sinclair, J.B. (1986) Colletotrichum destructivum, the anamorph of Glomerella glycines . Phytopathology, 76, 282–285. [Google Scholar]
- Manandhar, J.B. , Kunwar, K. , Tribhuwan Singh, G.L. , Hartman, G.L. & Sinclair, J.B. (1985) Penetration and infection of soybean leaf tissues by Colletotrichum truncatum and Glomerella glycine . Phytopathology, 75, 704–708. [Google Scholar]
- Mancini, V. , Murolo, S. & Romanazzi, G. (2016) Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathology, 65, 691–703. [Google Scholar]
- Marin‐Felix, Y. , Groenewald, J.Z. , Cai, L. , Chen, Q. , Marincowitz, S. , Barnes, I. et al. (2017) Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology, 86, 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B.A. (1997) The population genetics of fungi: Tools and techniques. Phytopathology, 87, 448–453. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. & Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology, 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Nagaraj, B.T. , Jahagirdar, S. & Basavaraja, G.T. (2014) Identification of resistant sources in glass house and field evaluation of soybean genotypes to anthracnose caused by Colletotrichum truncatum (Schew.) Andrus and Moore. The Bioscan, 9, 1333–1336. [Google Scholar]
- Nakata, K. & Takimoto, K. (1934) A list of crop diseases in Korea. In Agricultural experiment station governmental central chosen research report, 15, 1–146. [Google Scholar]
- Nataraj, V. , Maranna, S. , Kumawat, G. , Gupta, S. , Rajput, L.S. , Kumar, S. et al. (2020) Genetic inheritance and identification of germplasm sources for anthracnose resistance in soybean [Glycine max (L.) Merr.]. Genetic Resources and Crop Evolution, 67, 1449–1456. [Google Scholar]
- Nilsson, R.H. , Ryberg, M. , Kristiansson, E. , Abarenkov, K. , Larsson, K.‐H. & Kõljalg, U. (2006) Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS One, 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi, T. , Okayama, H. , Masubuchi, H. , Yonekawa, T. , Watanabe, K. , Amino, N. & Hase, T. (2000) Loop‐mediated isothermal amplification of DNA. Nucleic Acids Research, 28, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G. , Kleemann, J. , Torres, M.F. et al. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics, 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, R.P. & Hewitt, H.G. (2014) Fungicides in Crop Protection. Wallingford, UK: CABI. [Google Scholar]
- Oliver, R.P. & Solomon, P.S. (2010) New developments in pathogenicity and virulence of necrotrophs. Current Opinion in Plant Biology, 13, 415–419. [PubMed] [Google Scholar]
- Pagano, M.C. & Miransari, M. (2016) The importance of soybean production worldwide. In: Miransari, M. (Ed.) Abiotic and Biotic Stresses in Soybean Production. Cambridge, MA, USA: Academic Press, pp. 1–26. [Google Scholar]
- Pecchia, S. , Caggiano, B. , Da Lio, D. , Cafà, G. , Le Floch, G. & Baroncelli, R. (2019) Molecular detection of the seed‐borne pathogen Colletotrichum lupini targeting the hyper‐variable IGS region of the ribosomal cluster. Plants, 8, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino, C. , Gilardi, G. , Gullino, M.L. & Garibaldi, A. (2010) Detection of Phoma valerianellae in lambs lettuce seeds. Phytoparasitica, 38, 159–165. [Google Scholar]
- Perfect, S.E. , Hughes, H.B. , O’Connell, R.J. & Green, J.R. (1999) Colletotrichum: A model genus for studies on pathology and fungal–plant interactions. Fungal Genetics and Biology, 27, 186–198. [DOI] [PubMed] [Google Scholar]
- Pieck, M.L. , Ruck, A. , Farman, M.L. , Peterson, G.L. , Stack, J.P. , Valent, B. et al. (2017) Genomics‐based marker discovery and diagnostic assay development for wheat blast. Plant Disease, 101, 103–109. [DOI] [PubMed] [Google Scholar]
- Plissonneau, C. , Benevenuto, J. , Mohd‐Assaad, N. , Fouché, S. , Hartmann, F.E. & Croll, D. (2017) Using population and comparative genomics to understand the genetic basis of effector‐driven fungal pathogen evolution. Frontiers in Plant Science, 8, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poti, T. , Mahawan, K. , Cheewangkoon, R. , Arunothayanan, H. , Akimitsu, K. & Nalumpang, S. (2020) Detection and molecular characterization of carbendazim‐resistant Colletotrichum truncatum isolates causing anthracnose of soybean in Thailand. Journal of Phytopathology, 168, 267–278. [Google Scholar]
- Prasad, P. , Savadi, S. , Bhardwaj, S.C. , Gangwar, O.P. & Kumar, S. (2019) Rust pathogen effectors: Perspectives in resistance breeding. Planta, 250, 1–22. [DOI] [PubMed] [Google Scholar]
- Prusky, D. (1996) Pathogen quiescene in postharvest diseases. Annual Review of Phytopathology, 34, 413–434. [DOI] [PubMed] [Google Scholar]
- Prusky, D. , Alkan, N. , Mengiste, T. & Fluhr, R. (2013) Quiescent and necrotrophic lifestyle choice during postharvest disease development. Annual Review of Phytopathology, 51, 155–176. [Google Scholar]
- Raffaele, S. & Kamoun, S. (2012) Genome evolution in filamentous plant pathogens: Why bigger can be better. Nature Reviews Microbiology, 10, 417–430. [DOI] [PubMed] [Google Scholar]
- Rambaut, R. (2014) FigTree vol 1.4.2, a graphical viewer of phylogenetic trees. Available at: http://tree.bio.ed.ac.uk/software/figtree/ [Accessed 8 January 2021]. [Google Scholar]
- Ramiro, J. , Ciampi‐Guillardi, M. , Caldas, D.G.G. , de Moraes, M.H.D. , Barbieri, M.C.G. , Pereira, W.V. et al. (2019) Quick and accurate detection of Sclerotinia sclerotiorum and Phomopsis spp. in soybean seeds using qPCR and seed‐soaking method. Journal of Phytopathology, 167, 273–282. [Google Scholar]
- Ramos, A.M. , Gally, M. , García, M.C. & Levin, L. (2010) Pectinolytic enzyme production by Colletotrichum truncatum, causal agent of soybean anthracnose. Revista Iberoamericana De Micología, 27, 186–190. [DOI] [PubMed] [Google Scholar]
- Ramos, A.M. , Tadic, L.F. , Cinto, I. , Carmona, M. & Gally, M. (2013) Molecular characterization of Colletotrichum species causing soybean anthracnose in Argentina. Mycotaxon, 123, 457–465. [Google Scholar]
- Ranathunge, N.P. , Ford, R. & Taylor, P.W.J. (2009) Development and optimization of sequence‐tagged microsatellite site markers to detect genetic diversity within Colletotrichum capsici, a causal agent of chilli pepper anthracnose disease. Molecular Ecology Resources, 9, 1175–1179. [DOI] [PubMed] [Google Scholar]
- Rao, S. & Nandineni, M.R. (2017) Genome sequencing and comparative genomics reveal a repertoire of putative pathogenicity genes in chilli anthracnose fungus Colletotrichum truncatum . PLoS One, 12, e0183567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccioni, L. , Conca, G. & Hartman, G.L. (1998) First report of Colletotrichum coccodes on soybean in the United States. Plant Disease, 82, 959. [DOI] [PubMed] [Google Scholar]
- Rogério, F. , Boufleur, T.R. , Ciampi‐Guillardi, M. , Sukno, S.A. , Thon, M.R. , Massola Júnior, N.S. , et al. (2020) Genome sequence resources of Colletotrichum truncatum. C. plurivorum, C. musicola, and C. sojae: Four species pathogenic to soybean (Glycine max). Phytopathology, 110, 1497–1499. [DOI] [PubMed] [Google Scholar]
- Rogério, F. , Ciampi‐Guillardi, M. , Barbieri, M.C.G. , Bragança, C.A.D. , Seixas, C.D.S. , Almeida, A.M.R. et al. (2017) Phylogeny and variability of Colletotrichum truncatum associated with soybean anthracnose in Brazil. Journal of Applied Microbiology, 122, 402–415. [DOI] [PubMed] [Google Scholar]
- Rogério, F. , Gladieux, P. , Massola, N.S. & Ciampi‐Guillardi, M. (2019) Multiple introductions without admixture of Colletotrichum truncatum associated with soybean anthracnose in Brazil. Phytopathology, 109, 681–689. [DOI] [PubMed] [Google Scholar]
- Sant’Anna, J.R. , Miyamoto, C.T. , Rosada, L.J. , Franco, C.C.S. , Kaneshima, E.N. & Castro‐Prado, M.A.A. (2010) Genetic relatedness of Brazilian Colletotrichum truncatum isolates assessed by vegetative compatibility groups and RAPD analysis. Biological Research, 43, 51–62. [PubMed] [Google Scholar]
- Sarrocco, S. , Herrera‐Estrella, A. & Collinge, D.B. (2020) Editorial: Plant disease management in the post‐genomic era: From functional genomics to genome editing. Frontiers in Microbiology, 11, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, L. , Abdelfattah, A. , Mosca, S. , Li Destri Nicosia, M.G. , Agosteo, G.E. & Cacciola, S.O. (2017) Quantitative detection of Colletotrichum godetiae and C. acutatum sensu stricto in the phyllosphere and carposphere of olive during four phenological phases. European Journal of Plant Pathology, 149, 337–347. [Google Scholar]
- Sharma, R. (2009) Genetic differentiation of host limited forms of Colletotrichum truncatum from northwestern Himalayas. Archives of Phytopathology and Plant Protection, 42, 960–966. [Google Scholar]
- Sharma, S.K. , Gupta, G.K. & Ramteke, R. (2011) Colletotrichum truncatum [(Schw.) Andrus and W.D. Moore], the causal agent of anthracnose of soybean [Glycine max (L.) Merrill.] – a review. Soybean Research, 9, 31–52. [Google Scholar]
- Shi, X. , Wang, S. , Duan, X. , Gao, X. , Zhu, X. & Laborda, P. (2020) First report of Colletotrichum brevisporum causing soybean antrhacnose in China. Plant Disease, 10.1094/PDIS-09-20-1910-PDN [DOI] [PubMed] [Google Scholar]
- de Silva, D.D. , Crous, P.W. , Ades, P.K. , Hyde, K.D. & Taylor, P.W.J. (2017) Lifestyles of Colletotrichum species and implications for plant biosecurity. Fungal Biology Reviews, 31, 155–168. [Google Scholar]
- da Silva, L.L. , Moreno, H.L.A. , Correia, H.L.N. , Santana, M.F. & de Queiroz, M.V. (2020) Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Applied Microbiology and Biotechnology, 104, 1891–1904. [DOI] [PubMed] [Google Scholar]
- Sinclair, J.B. (1989) Compendium of Soybean Diseases, 3rd edition. St Paul, MN, USA: American Phytopathological Society. [Google Scholar]
- Stecher, G. , Tamura, K. & Kumar, S. (2020) Molecular evolutionary genetics analysis (MEGA) for macOS. Molecular Biology and Evolution, 37, 1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E.H. (2016) The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology, 106, 104–112. [DOI] [PubMed] [Google Scholar]
- Stukenbrock, E.H. & Bataillon, T. (2012) A population genomics perspective on the emergence and adaptation of new plant pathogens in agro‐ecosystems. PLoS Pathogens, 8, e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi, S. , Gharti, D.B. , Neupane, S. & Ghimire, T. (2016) Management of anthracnose in soybean using fungicide. Journal of Nepal Agricultural Research Council, 1, 29–32. [Google Scholar]
- Talhinhas, P. , Baroncelli, R. & Le Floch, G. (2016) Anthracnose of lupins caused by Colletotrichum lupini: A recent disease and a successful worldwide pathogen. Journal of Plant Pathology, 98, 5–14. [Google Scholar]
- Thierry, M. , Gladieux, P. , Fournier, E. , Tharreau, D. & Ioos, R. (2020) A genomic approach to develop a new qPCR test enabling detection of the Pyricularia oryzae lineage causing wheat blast. Plant Disease, 104, 60–70. [DOI] [PubMed] [Google Scholar]
- Tian, Q. , Lu, C. , Wang, S. , Xiong, Q. , Zhang, H. , Wang, Y. & et al (2017) Rapid diagnosis of soybean anthracnose caused by Colletotrichum truncatum using a loop‐mediated isothermal amplification (LAMP) assay. European Journal of Plant Pathology, 148, 785–793. [Google Scholar]
- Ureña‐Padilla, A.R. , MacKenzie, S.J. , Bowen, B.W. & Legard, D.E. (2002) Etiology and population genetics of Colletotrichum spp. causing crown and fruit rot of strawberry. Phytopathology, 92, 1245–1252. [DOI] [PubMed] [Google Scholar]
- USDA (2020) World Agricultural Production. Circular Series WAP 12‐20. USA: USDA. https://downloads.usda.library.cornell.edu/usda‐esmis/files/5q47rn72z/cr56ns297/00000r529/production.pdf [Google Scholar]
- Van de Wouw, A.P. & Idnurm, A. (2019) Biotechnological potential of engineering pathogen effector proteins for use in plant disease management. Biotechnology Advances, 37, 107387. [DOI] [PubMed] [Google Scholar]
- Vasconcelos, M.J.V. , Machado, M.A. , Almeida, A.M.D. , Henning, A.A. , de Barros, E.G. & Moreira, M.A. (1994) Differentiation of Colletotrichum truncatum isolates by random amplified polymorphic DNA. Fitopatologia Brasileira, 19, 520–523. [Google Scholar]
- Vieira, W.A.D.S. , Bezerra, P.A. , Silva, A.C.D. , Veloso, J.S. , Câmara, M.P.S. & Doyle, V.P. (2020) Optimal markers for the identification of Colletotrichum species. Molecular Phylogenetics and Evolution, 143, 106694. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Ye, W. , Tian, Q. , Dong, S. & Zheng, X. (2017) Rapid detection of Colletotrichum gloeosporioides using a loop‐mediated isothermal amplification assay. Australasian Plant Pathology, 46, 493–498. [Google Scholar]
- Weir, B.S. , Johnston, P.R. & Damm, U. (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology, 73, 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrather, A. , Shannon, G. , Balardin, R. , Carregal, L. , Escobar, R. , Gupta, G.K. et al. (2010) Effect of diseases on soybean yield in the top eight producing countries in 2006. Plant Health Progress, 11, 29. [Google Scholar]
- Xavier, K.V. , Mizubuti, E.S.G. , Queiroz, M.V. , Chopra, S. & Vaillancourt, L. (2018) Genotypic and pathogenic diversity of Colletotrichum sublineola isolates from sorghum (Sorghum bicolor) and johnsongrass (S. halepense) in the southeastern United States. Plant Disease, 102, 2341–2351. [DOI] [PubMed] [Google Scholar]
- Yang, H.‐C. & Hartman, G.L. (2015) Methods and evaluation of soybean genotypes for resistance to Colletotrichum truncatum . Plant Disease, 99, 143–148. [DOI] [PubMed] [Google Scholar]
- Yang, H.C. & Hartman, G.L. (2016) Anthracnose. In: Hartman, G.L. , Rupe, J. and Sikora, E.J. (Eds.) Compendium of Soybean Diseases and Pests. St Paul, MN, USA: American Phytopathological Society, pp. 31–34. [Google Scholar]
- Yang, H.‐C. , Haudenshield, J.S. & Hartman, G.L. (2012) First report of Colletotrichum chlorophyti causing soybean anthracnose. Plant Disease, 96, 1699. [DOI] [PubMed] [Google Scholar]
- Yang, H.‐C. , Haudenshield, J.S. & Hartman, G.L. (2014) Colletotrichum incanum sp. nov., a curved‐conidial species causing soybean anthracnose in USA. Mycologia, 106, 32–42. [DOI] [PubMed] [Google Scholar]
- Yang, H.‐C. , Haudenshield, J.S. & Hartman, G.L. (2015) Multiplex real‐time PCR detection and differentiation of Colletotrichum species infecting soybean. Plant Disease, 99, 1559–1568. [DOI] [PubMed] [Google Scholar]
- Yang, H.‐C. , Stewart, J.M. & Hartman, G.L. (2013) First report of Colletotrichum chlorophyti infecting soybean seed in Arkansas, United States. Plant Disease, 97, 1510. [DOI] [PubMed] [Google Scholar]
- Zaw, M. , Aye, S.S. & Matsumoto, M. (2020) Colletotrichum and Diaporthe species associated with soybean stem diseases in Myanmar. Journal of General Plant Pathology, 86, 114–123. [Google Scholar]
- Zhan, J. (2009) Population genetics of plant pathogens. In: Encyclopedia of Life Sciences. John Wiley and Sons Ltd, a0021269. https://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0021269 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Strains of Colletotrichum spp. studied, with collection details and GenBank accession numbers
Data Availability Statement
Data sharing is not applicable as no new data were generated or analysed.


