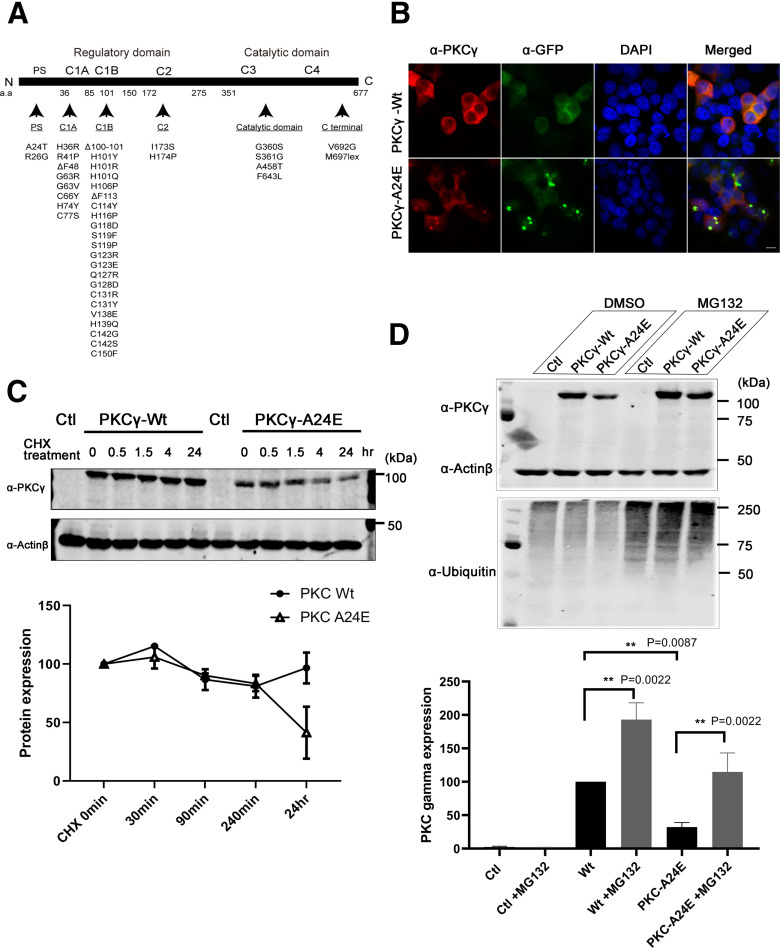
Figure 1.
Pseudosubstrate mutant PKCγ protein is unstable and shows aggregation. A, Illustrations of PKCγ protein domain mutations and deletions found in SCA14 families. Most mutations are found in the C1B domain. B, The 5 µg of GFP-Control plasmid, GFP-PKCγ-Wt, or GFP-PKCγ-A24E was transfected to the HeLa cells; and after 24 h, cells were fixed with PFA following the immunostaining. GFP-PKCγ-A24E showed aggregation and accumulated in HELA cells. Images were acquired with confocal microscopy (Carl Zeiss, LSM700) using a Plan-Apochromat 100×/1.3 Oil DIC M27 objective (Carl Zeiss). Scale bar, 10 µm. C, Pseudosubstrate domain mutant PKCγ is unstable and is degraded after 35 µg/ml cycloheximide treatment. After 48 h transfection, cycloheximide was applied to the cells. Samples were collected at 0 min, 30 min, 90 min, 240 min, and 24 h. Twenty-four hours after cycloheximide treatment, the PKCγ protein expression level is GFP-PKCγ-Wt = 96.68% and GFP-PKCγ-A24E = 41.27% compared with each starting point, respectively. D, Degradation of pseudosubstrate domain mutant PKCγ occurs via the proteasome pathway. Twenty-four hours after 5 μm of proteasome inhibitor MG132 treatment, HEK293T cells show more ubiquitinated proteins, and this treatment rescued GFP-PKCγ-A24E protein levels (GFP-PKCγ-Wt = 100.0%; GFP-PKCγ-Wt + MG132 = 192.90% ± 25.35; GFP-PKCγ-A24E = 32.26% ± 6.75; GFP-PKCγ-A24E + MG132 = 114.60% ± 28.54; n = 6). Protein expression was analyzed using the two-tailed Mann–Whitney test (GFP-PKCγ-Wt vs GFP-PKCγ-A24E, p = 0.0022, GFP-PKCγ-Wt vs GFP-PKCγ-Wt + MG132, p = 0.0022; GFP-PKCγ-A24E vs GFP-PKCγ-A24E + MG132, p = 0.0087).