Figure 2.
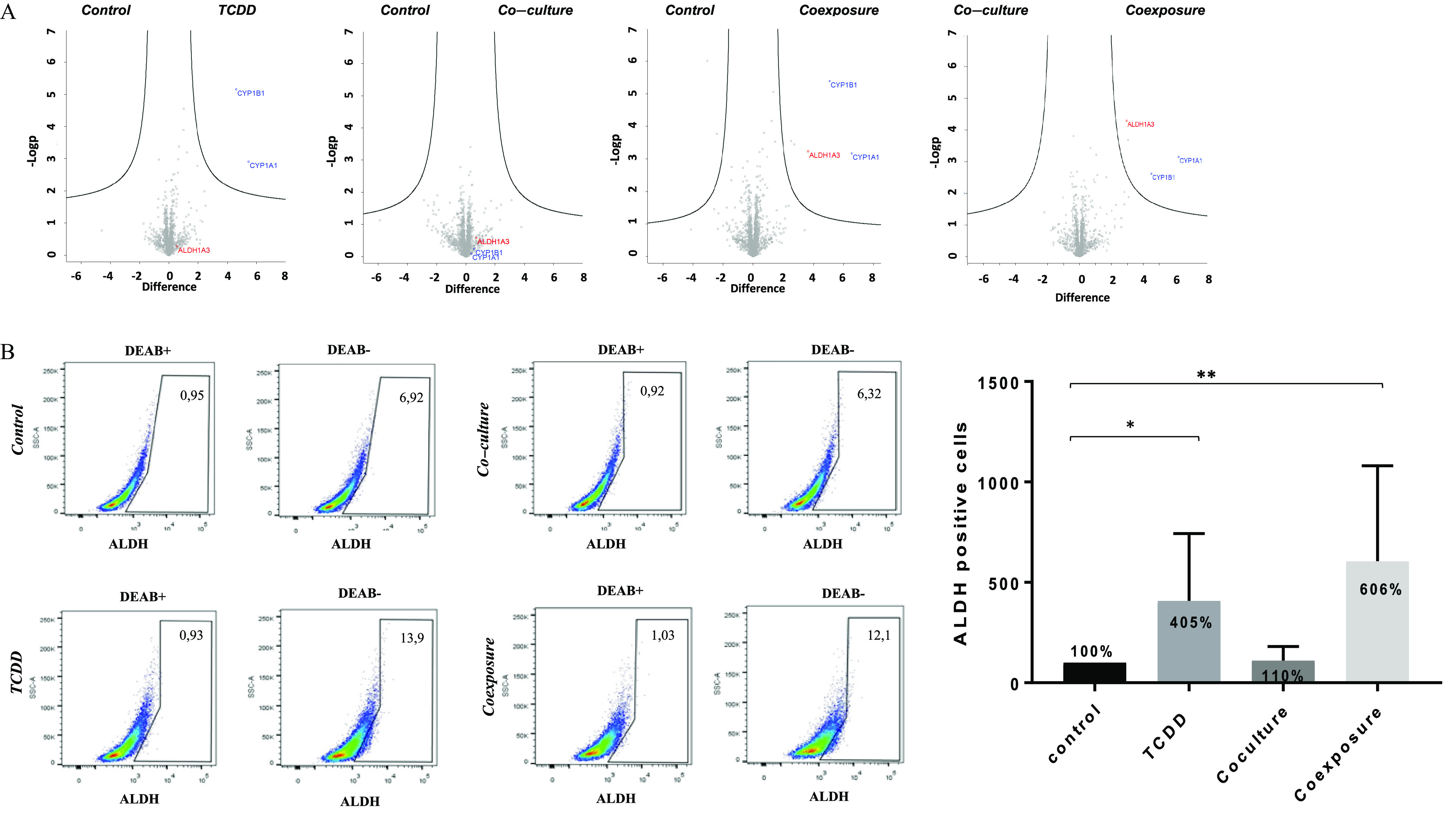
Proteomics analysis and ALDH enzymatic analysis of MCF7 cells growth in co-culture with hMADS and exposed to TCDD for 48 h. (A) High-throughput proteomic analysis of MCF7 cells [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS), and coexposure (co-culture with TCDD)]. The plots show the mean of biological quadruplicates and technical triplicates for each sample. CYP1A1, CYP1B1, and ALDH1A3 were induced when they appeared in the upper right part of the representation. (B) ALDH (aldehyde dehydrogenase) enzymatic activity was detected in MCF7 cells using the ALDEFLUOR assay (FACS analysis). DEAB was used to inhibit the reaction of ALDH with the ALDEFLUOR reagent, providing a negative control. Percentage of ALDH positive cell was set according to the gate of DEAB control cells. Graph represents means of the percentage of ALDH positive cells (in bold) compared with the of six measurements. The numerical information and p-values are provided in Table S2. (Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, **; *). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
